Import wizard for tab separated lists
With this import routine, you can import data from text
files (as tab-separated lists) into the database. Choose Data ->
 Import ->
Import ->
 Import wizard and then the type of data that should be imported, e.g.
Import wizard and then the type of data that should be imported, e.g.
 Reference import ... from the menu. A window as shown below will open
that will lead you through the import of the data. The window is separated in 3
areas. On the left side, you see a list of possible data related import steps according
to the type of data you choose for the import. On the right side you see the list
of currently selected import steps. In the middle part the details of the selected
import steps are shown.
Reference import ... from the menu. A window as shown below will open
that will lead you through the import of the data. The window is separated in 3
areas. On the left side, you see a list of possible data related import steps according
to the type of data you choose for the import. On the right side you see the list
of currently selected import steps. In the middle part the details of the selected
import steps are shown.
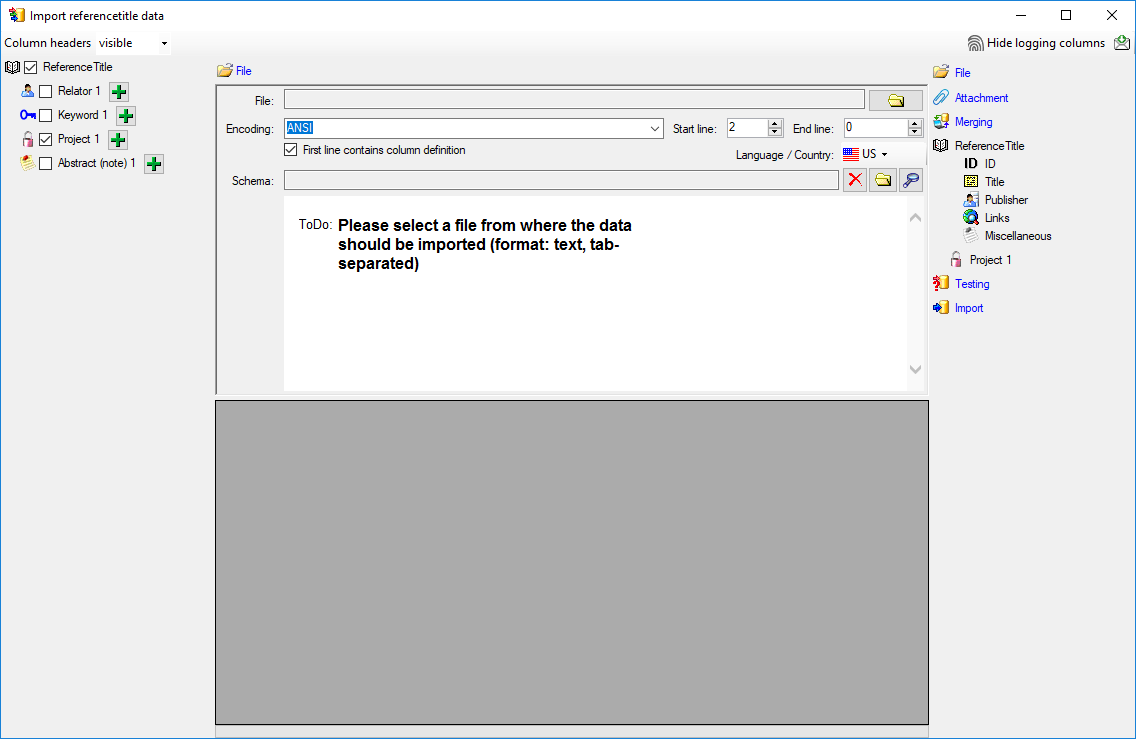
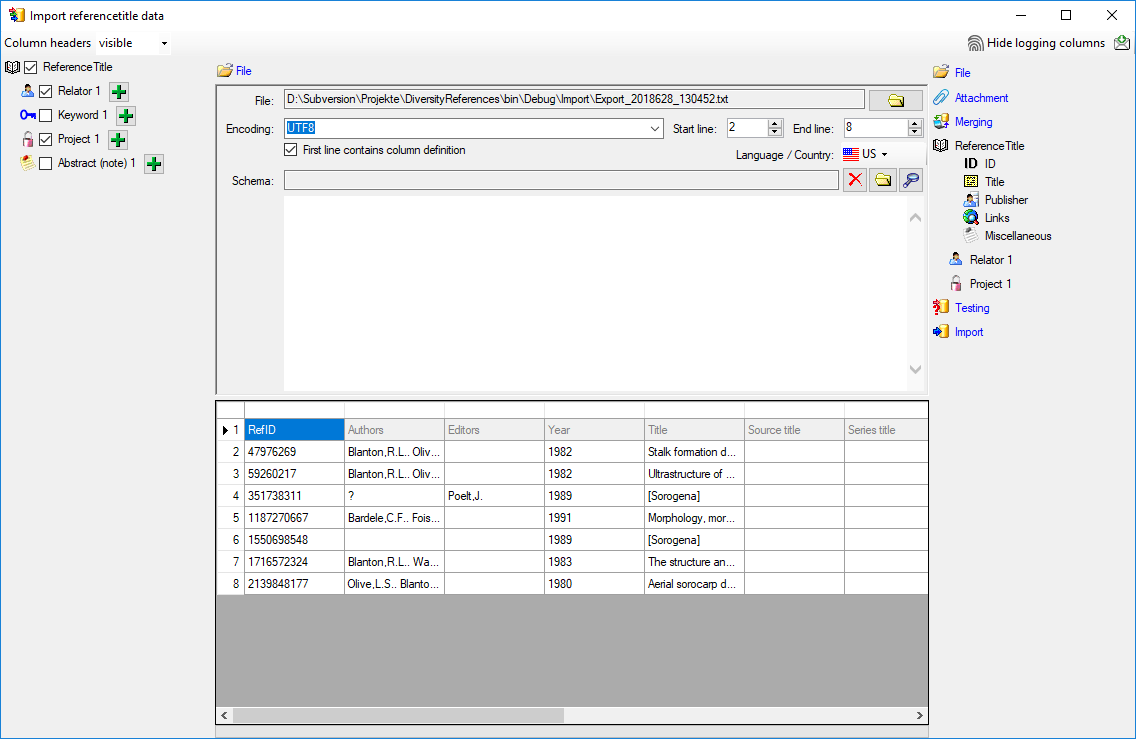
As a first step, choose the
 File from where the data should
be imported. The currently supported format is tab-separated text.
Then choose the Encoding of the file, e.g. Unicode. The preferred encoding
is UTF8. The Start line and End line will automatically be set according
to your data. You may change these to restrict the data lines that should be imported.
The not imported parts
in the file are indicated as shown below with a gray background. If the
File from where the data should
be imported. The currently supported format is tab-separated text.
Then choose the Encoding of the file, e.g. Unicode. The preferred encoding
is UTF8. The Start line and End line will automatically be set according
to your data. You may change these to restrict the data lines that should be imported.
The not imported parts
in the file are indicated as shown below with a gray background. If the
 First line contains the column definition this line will not be imported
as well. If your data contains e.g. date information where notations differ between
countries (e.g. 31.4.2013 - 4.31.2013), choose the Language / Country to
ensure a correct interpretation of your data. Finally you can select a prepared
Schema (see chapter Schema below) for the import.
First line contains the column definition this line will not be imported
as well. If your data contains e.g. date information where notations differ between
countries (e.g. 31.4.2013 - 4.31.2013), choose the Language / Country to
ensure a correct interpretation of your data. Finally you can select a prepared
Schema (see chapter Schema below) for the import.
Choosing the data ranges
In the selection list on the left side of the window (see
below) all possible import steps for the data are listed according to the type of
data you want to import.
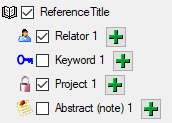
The import of certain tables can be paralleled. To add
parallels click on the
 button (see below). To remove parallels, use the
button (see below). To remove parallels, use the
 button. Only selected ranges will appear in the list of the steps on the right (see
below).
button. Only selected ranges will appear in the list of the steps on the right (see
below).

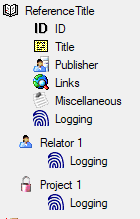
To import information of logging columns like who created
and changed the data, click on
 button in the header line. This will include an additional substeps for every step
containing the logging columns (see below). If you do not import these data, they
will be automatically filled by default values like the current time and user.
button in the header line. This will include an additional substeps for every step
containing the logging columns (see below). If you do not import these data, they
will be automatically filled by default values like the current time and user.
You can either import your data as new data or
 Attach
them to data in the database. Select the import step
Attach
them to data in the database. Select the import step
 Attachment
from the list. All tables that are selected and contain columns at which you can
attach data are listed (see below). Either choose the first option
Attachment
from the list. All tables that are selected and contain columns at which you can
attach data are listed (see below). Either choose the first option
 Import as new data or one of the columns the attachment columns offered like
SeriesCode in the table Series in the example below.
Import as new data or one of the columns the attachment columns offered like
SeriesCode in the table Series in the example below.

If you select a column for attachment, this column will
be marked with a blue background (see below and chapter Table data).

You can either import your data as new data or
 Merge them with data in the database. Select the import step
Merge them with data in the database. Select the import step
 Merge from the list. For every table you can choose between
Merge from the list. For every table you can choose between
 Insert, img alt="" src="img/IcoMergeMerge.png" /> Merge,
Insert, img alt="" src="img/IcoMergeMerge.png" /> Merge,
 Update and
Update and
 Attach (see below).
Attach (see below).
The
 Insert option will import the data from the file independent of existing data
in the database.
Insert option will import the data from the file independent of existing data
in the database.
The
 Merge option will compare the data from the file with those in the database
according to the
Merge option will compare the data from the file with those in the database
according to the
 Key columns (see below). If no matching data are found in the database, the
data from the file will be imported. Otherwise the data will be updated.
Key columns (see below). If no matching data are found in the database, the
data from the file will be imported. Otherwise the data will be updated.
The
 Update option will compare the data from the file with those in the database
according to the
Update option will compare the data from the file with those in the database
according to the
 Key columns. Only matching data found in the database will be updated.
Key columns. Only matching data found in the database will be updated.
The
 Attach option will compare the data from the file with those in the database
according to the
Attach option will compare the data from the file with those in the database
according to the
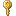 Key columns. The found data will not be changed, but used as a reference
data in depending tables.
Key columns. The found data will not be changed, but used as a reference
data in depending tables.
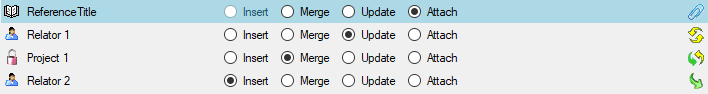
Table data
To set the source for the columns in the file, select
the step of a table listed underneath the
 Merge step. All columns available for importing data will be listed in the
central part of the window. In the example shown below, the first column is used
to attach the new data to data in the database.
Merge step. All columns available for importing data will be listed in the
central part of the window. In the example shown below, the first column is used
to attach the new data to data in the database.
A reminder in the header line will show you what actions
are still needed to import the data into the table:
- Please select at least one column
 = No column
has been selected so far.
= No column
has been selected so far.
- Please select at least one decisive column
 = If data
will be imported depends on the content of decisive columns, so at least one must
be selected.
= If data
will be imported depends on the content of decisive columns, so at least one must
be selected.
- Please select the position in the file
 = The
position in the file must be given if the data for a column should be taken from
the file.
= The
position in the file must be given if the data for a column should be taken from
the file.
- Please select at least one column for
comparison
 = For
all merge types other than insert columns for comparison with data in the database
are needed.
= For
all merge types other than insert columns for comparison with data in the database
are needed.
- From file or For all
 = For
every you have to decide whether the data are taken from the file or a value is
entered for all
= For
every you have to decide whether the data are taken from the file or a value is
entered for all
- Please select a value from the list
 = You have
to select a value from the provided list
= You have
to select a value from the provided list
- Please enter a value
 = You
have to enter a value used for all datasets
= You
have to enter a value used for all datasets
The handling of the columns in described in the chapter
columns.
To test if all requirements for the import are met use
the
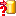 Testing step. You can use a certain line in the file for you test and then click
on the Test data in line: button. If there are still unmet requirements,
these will be listed in a window as shown below.
Testing step. You can use a certain line in the file for you test and then click
on the Test data in line: button. If there are still unmet requirements,
these will be listed in a window as shown below.
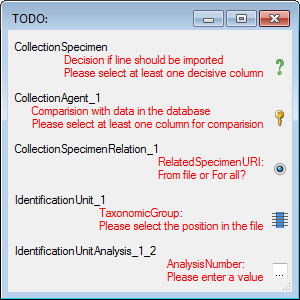
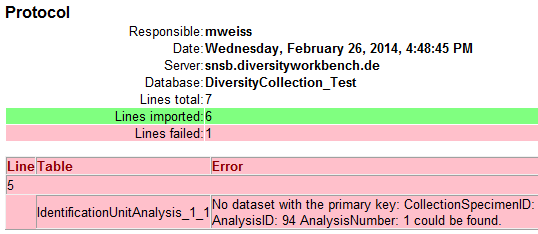
If finally all requirements are met, the testing function
will try to write the data into the database and display any errors that occurred
as shown below. All datasets marked with a red
background, produced some error.
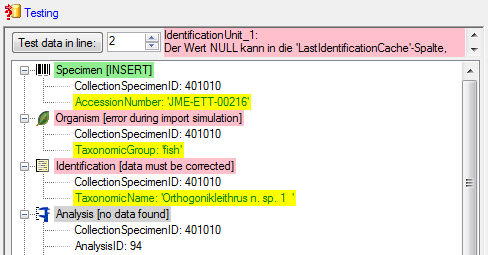
To see the list of all errors, double click in the error list window in the header line (see
below).
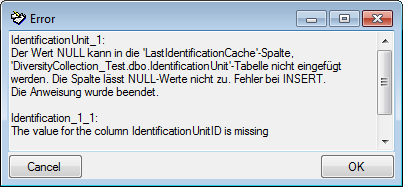
If finally no errors are left, your data are ready for
import. The colors in the table nodes in the tree indicate the handling of the datasets:
INSERT,
MERGE, UPDATE,
No difference. Attach,
No data. The colors of the table
columns indicate whether a column is decisive
 , a
key column
, a
key column
 or an attachment column
or an attachment column
 .
.
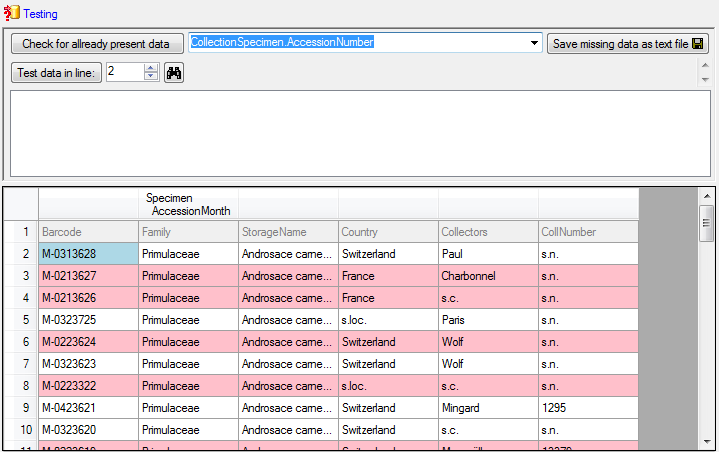
If you suspect, that the import file contains data already
present in the database, you may test this and extract only the missing lines in
a new file. Choose the attachment column (see chapter Attaching data) and click
on the button Check for already present data. The data already present in
the database will be marked red (see
below). Click on the button Save missing data as text file
 to store the data not present in the database in a new file for the import. The
import of specimen contains the option
to store the data not present in the database in a new file for the import. The
import of specimen contains the option
 Use default duplicate check for AccessionNumber that is selected by default.
Use default duplicate check for AccessionNumber that is selected by default.
If you happen to get a file with a content as shown below,
you may have seleted the wrong encoding or the encoding is incompatible. Please
try to save the original file as UTF8 and select this encoding for the import.

With the last step you can finally start to import the
data into the database. If you want to repeat the import with the same settings
and data of the same structure, you can save a schema of the current settings (see
below). You optionally can include a description of you schema and with the
 button you can generate a file containing
only the description.
button you can generate a file containing
only the description.
|
Schedule for import of tab-separated text files into DiversityCollection
|
|
Target within DiversityCollection: Specimen
|
|
Schedule version:
|
1
|
Database version:
|
02.05.41
|
|
Lines:
|
2 - 3215
|
First line contains column definition:
|
?
|
|
Encoding:
|
Unicode
|
Language:
|
de
|
Tables
CollectionSpecimen (CollectionSpecimen)
Parent: CollectionEvent
Merge handling:
Insert
|
Column in table
|
?
|
Key
|
Copy
|
Pre
|
Post
|
File pos.
|
Transformations
|
Value
|
Source
|
Table
|
|
CollectionSpecimenID
|
|
|
|
|
|
|
|
|
Database
|
|
|
AccessionNumber
|
?
|
?
|
|
|
|
0
|
|
|
File
|
|
IdentificationUnit_1 (IdentificationUnit)
Parent: CollectionSpecimen
Merge handling:
Merge
|
Column in table
|
?
|
Key
|
Copy
|
Pre
|
Post
|
File pos.
|
Transformations
|
Value
|
Source
|
Table
|
|
CollectionSpecimenID
|
|
|
|
|
|
|
|
|
Database
|
|
|
IdentificationUnitID
|
|
|
|
|
|
|
|
|
Database
|
|
|
LastIdentificationCache
|
|
?
|
|
|
|
2
|
|
|
File
|
|
|
+
|
|
|
|
|
|
3
|
|
|
File
|
|
|
+
|
|
|
|
|
|
4
|
|
|
File
|
|
|
+
|
|
|
|
|
|
5
|
|
|
File
|
|
|
TaxonomicGroup
|
?
|
|
|
|
|
|
|
fish
|
Interface
|
|
IdentificationUnitAnalysis_1_1
(IdentificationUnitAnalysis)
Parent: IdentificationUnit_1
Merge handling:
Update
|
Column in table
|
?
|
Key
|
Copy
|
Pre
|
Post
|
File pos.
|
Transformations
|
Value
|
Source
|
Table
|
|
CollectionSpecimenID
|
|
|
|
|
|
|
|
|
Database
|
|
|
IdentificationUnitID
|
|
|
|
|
|
|
|
|
Database
|
|
|
AnalysisID
|
|
|
|
|
|
|
|
94
|
Interface
|
|
|
AnalysisNumber
|
|
|
|
|
|
|
|
1
|
Interface
|
|
|
AnalysisResult
|
?
|
?
|
|
|
|
39
|
|
|
File
|
|
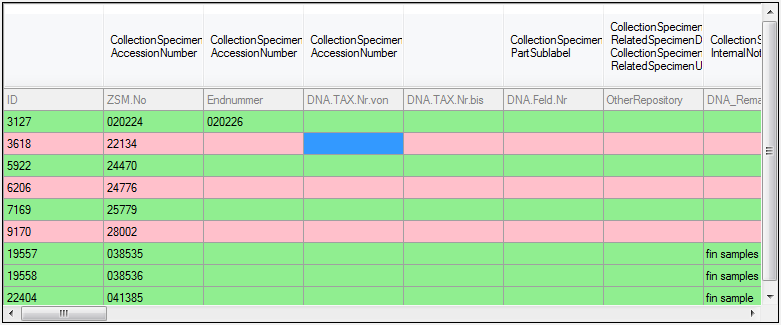
Lines that could not be imported will be marked with a
red background while imported lines are marked green (see below).
If you want to save lines that produce errors during the
import in a separate file, use the Save failed lines option. The protocol of the
import will contain all settings according to the used schema and an overview containing
the number of inserted, updated, unchanged and failed lines (see below).
Description

A description of the schema may be included in the schema
itself or with a click on the
 button generated as a separate file.
This file will be located in a separate directory Description to avoid confusion
with import schemas. An example for a description file is shown below, containing
common settings, the treatment of the file columns and interface settings as defined
in the schema.
button generated as a separate file.
This file will be located in a separate directory Description to avoid confusion
with import schemas. An example for a description file is shown below, containing
common settings, the treatment of the file columns and interface settings as defined
in the schema.
|
Schedule for import of tab-separated text files into DiversityCollection
|
|
Target within DiversityCollection: Specimen
|
|
Schedule version:
|
1
|
Database version:
|
02.05.52
|
|
Lines:
|
2 - 5
|
First line contains column definition:
|
?
|
|
Encoding:
|
Unicode
|
Language:
|
de
|
|
Description:
|
Import Schema fuer Literaturdaten (Bayernflora) aus Dörr & Lippert mit MTB Daten
und max. 4 Sammlern
|
File columns
|
Merge handling of table
|
Column usage
|
|
|
|
Insert
|
Merge
|
Update
|
Attach
|
Decisive
|
Key
|
|
|
|
Pos.
|
Name
|
Table
|
Column
|
Example
|
Transformed
|
|
0
|
ID
|
|
CollectionSpecimen.
|
ExternalIdentifier
|
1
|
|
|
1
|
originalname
|
|
Identification_1_2.
|
TaxonomicName
|
Ophioglossum vulgatum
|
|
|
2
|
nameautor
|
|
Identification_1_1.
|
TaxonomicName
|
Ophioglossum vulgatum L.
|
|
|
3
|
taxnr
|
|
Identification_1_1.
|
NameURI
|
3949
|
|
|
|
|
Prefix:
|
http://tnt.diversityworkbench.de/TaxonNames_Plants/
|
|
4
|
mtb
|
|
CollectionEventLocalisation_6.
|
Location1
|
8423
|
|
|
5
|
qu
|
|
CollectionEventLocalisation_6.
|
Location2
|
2
|
|
|
6
|
unschärfe
|
|
CollectionEventLocalisation_6.
|
LocationAccuracy
|
|
|
|
7
|
jahr_von
|
|
CollectionEvent.
|
CollectionYear
|
1902
|
|
|
8
|
jahr_bis
|
|
CollectionEvent.
|
CollectionDateSupplement
|
|
|
|
|
|
Prefix:
|
bis?
|
|
9
|
status
|
|
IdentificationUnitAnalysis_1_1.
|
AnalysisResult
|
|
|
|
10
|
verwaltungseinheit
|
not imported
|
|
11
|
fundort
|
not imported
|
|
12
|
finder
|
not imported
|
|
13
|
ID_collector1
|
|
CollectionAgent_1.
|
CollectorsAgentURI
|
43708
|
|
|
|
|
Prefix:
|
http://snsb.diversityworkbench.de/Agents_BayernFlora/
|
|
14
|
ID_collector2
|
|
CollectionAgent_2.
|
CollectorsAgentURI
|
|
|
|
|
|
Prefix:
|
http://snsb.diversityworkbench.de/Agents_BayernFlora/
|
|
15
|
ID_collector3
|
|
CollectionAgent_3.
|
CollectorsAgentURI
|
|
|
|
|
|
Prefix:
|
http://snsb.diversityworkbench.de/Agents_BayernFlora/
|
|
16
|
ID_collector4
|
|
CollectionAgent_4.
|
CollectorsAgentURI
|
|
|
|
|
|
Prefix:
|
http://snsb.diversityworkbench.de/Agents_BayernFlora/
|
|
17
|
primärquelle
|
not imported
|
|
18
|
ID_primärquelle
|
|
Annotation_1.
|
ReferenceURI
|
|
|
|
|
|
Prefix:
|
http://id.snsb.info/references/
|
|
19
|
primärquelle_seiten
|
not imported
|
|
20
|
bestand
|
|
IdentificationUnitAnalysis_1_2.
|
AnalysisResult
|
|
|
|
21
|
sonstiges
|
|
CollectionSpecimen.
|
OriginalNotes
|
|
|
|
22
|
höhe
|
|
CollectionEventLocalisation_7.
|
Location1
|
|
|
|
23
|
herbar1
|
not imported
|
|
24
|
herbar2
|
not imported
|
|
25
|
herbar3
|
not imported
|
|
26
|
ID_herbar1
|
|
CollectionSpecimenRelation_1.
|
RelatedSpecimenCollectionID
|
|
|
|
27
|
ID_herbar2
|
not imported
|
|
28
|
ID_herbar3
|
not imported
|
|
29
|
det
|
not imported
|
|
30
|
ID_det
|
not imported
|
|
31
|
rev
|
not imported
|
|
32
|
ID_rev
|
not imported
|
|
33
|
datenquelle
|
not imported
|
|
34
|
ID_datenquelle
|
|
CollectionSpecimen.
|
ReferenceURI
|
135
|
|
|
|
|
Prefix:
|
http://id.snsb.info/references/
|
|
35
|
project1
|
not imported
|
|
36
|
project2
|
|
CollectionSpecimen.
|
AdditionalNotes
|
O
|
Beobachtung
|
|
|
|
Transformations:
|
|
Reglar express.:
|
|
O
|
?
|
Beobachtung
|
|
|
Reglar express.:
|
|
H
|
?
|
Herbarauswertung
|
|
|
Reglar express.:
|
|
L
|
?
|
Literaturauswertung
|
|
|
Interface settings
|
Table
|
Table alias
|
Column
|
Value
|
|
Annotation
|
Annotation_1
|
AnnotationType
|
Reference
|
|
Annotation_1
|
Annotation
|
Literaturauswertung: nach Dörr & Lippert (2004)
|
|
Annotation_1
|
ReferenceDisplayText
|
Annotation
|
|
CollectionAgent
|
CollectionAgent_1
|
CollectorsName
|
Collector1
|
|
CollectionAgent_2
|
CollectorsName
|
Collector2
|
|
CollectionAgent_3
|
CollectorsName
|
Collector3
|
|
CollectionAgent_4
|
CollectorsName
|
Collector4
|
|
CollectionEvent
|
|
CountryCache
|
Germany
|
|
CollectionProject
|
CollectionProject_1
|
ProjectID
|
37
|
|
CollectionProject_2
|
ProjectID
|
149
|
|
CollectionSpecimen
|
|
ReferenceTitle
|
Reference
|
|
CollectionSpecimenRelation
|
CollectionSpecimenRelation_1
|
RelatedSpecimenURI
|
|
|
CollectionSpecimenRelation_1
|
RelatedSpecimenDisplayText
|
|
|
CollectionSpecimenRelation_1
|
Notes
|
Herbarauswertung: nach Dörr & Lippert (2004)
|
|
Identification
|
Identification_1_1
|
IdentificationSequence
|
2
|
|
Identification_1_2
|
IdentificationSequence
|
1
|
|
Identification_1_2
|
Notes
|
Originalname aus Dörr & Lippert (2004)
|
|
IdentificationUnit
|
IdentificationUnit_1
|
LastIdentificationCache
|
plant
|
|
IdentificationUnit_1
|
TaxonomicGroup
|
plant
|
|
IdentificationUnitAnalysis
|
IdentificationUnitAnalysis_1_1
|
AnalysisID
|
2
|
|
IdentificationUnitAnalysis_1_1
|
AnalysisNumber
|
1
|
|
IdentificationUnitAnalysis_1_2
|
AnalysisID
|
4
|
|
IdentificationUnitAnalysis_1_2
|
AnalysisNumber
|
2
|
 Import ->
Import ->
 Import wizard and then the type of data that should be imported, e.g.
Import wizard and then the type of data that should be imported, e.g.
 Reference import ... from the menu. A window as shown below will open
that will lead you through the import of the data. The window is separated in 3
areas. On the left side, you see a list of possible data related import steps according
to the type of data you choose for the import. On the right side you see the list
of currently selected import steps. In the middle part the details of the selected
import steps are shown.
Reference import ... from the menu. A window as shown below will open
that will lead you through the import of the data. The window is separated in 3
areas. On the left side, you see a list of possible data related import steps according
to the type of data you choose for the import. On the right side you see the list
of currently selected import steps. In the middle part the details of the selected
import steps are shown.


 First line contains the column definition this line will not be imported
as well. If your data contains e.g. date information where notations differ between
countries (e.g. 31.4.2013 - 4.31.2013), choose the Language / Country to
ensure a correct interpretation of your data. Finally you can select a prepared
Schema (see chapter Schema below) for the import.
First line contains the column definition this line will not be imported
as well. If your data contains e.g. date information where notations differ between
countries (e.g. 31.4.2013 - 4.31.2013), choose the Language / Country to
ensure a correct interpretation of your data. Finally you can select a prepared
Schema (see chapter Schema below) for the import.


 button (see below). To remove parallels, use the
button (see below). To remove parallels, use the
 button. Only selected ranges will appear in the list of the steps on the right (see
below).
button. Only selected ranges will appear in the list of the steps on the right (see
below).
 button in the header line. This will include an additional substeps for every step
containing the logging columns (see below). If you do not import these data, they
will be automatically filled by default values like the current time and user.
button in the header line. This will include an additional substeps for every step
containing the logging columns (see below). If you do not import these data, they
will be automatically filled by default values like the current time and user.

 Import as new data or one of the columns the attachment columns offered like
SeriesCode in the table Series in the example below.
Import as new data or one of the columns the attachment columns offered like
SeriesCode in the table Series in the example below.



 Insert, img alt="" src="img/IcoMergeMerge.png" /> Merge,
Insert, img alt="" src="img/IcoMergeMerge.png" /> Merge,
 Update and
Update and
 Key columns (see below). If no matching data are found in the database, the
data from the file will be imported. Otherwise the data will be updated.
Key columns (see below). If no matching data are found in the database, the
data from the file will be imported. Otherwise the data will be updated.


 = If data
will be imported depends on the content of decisive columns, so at least one must
be selected.
= If data
will be imported depends on the content of decisive columns, so at least one must
be selected. = The
position in the file must be given if the data for a column should be taken from
the file.
= The
position in the file must be given if the data for a column should be taken from
the file.  = You have
to select a value from the provided list
= You have
to select a value from the provided list  = You
have to enter a value used for all datasets
= You
have to enter a value used for all datasets 



 to store the data not present in the database in a new file for the import. The
import of specimen contains the option
to store the data not present in the database in a new file for the import. The
import of specimen contains the option




