Subsections of Synchronizing
Maintenance
Gazetteer
Maintenance - Gazetteer
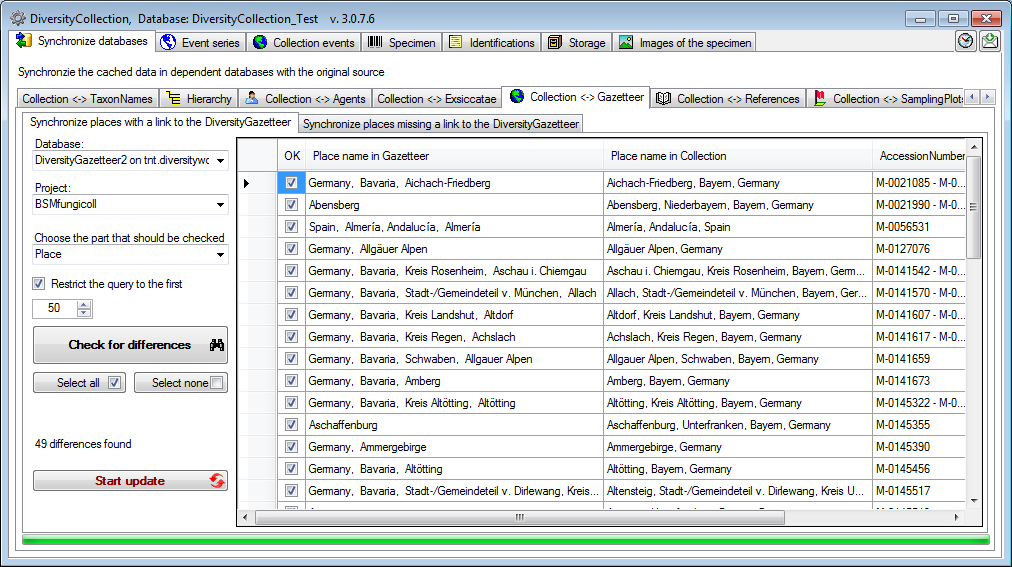
To synchronize the entries derived from the module DiversityGazetteer
choose Administration → Maintenance from the menu. A window will
open as shown below. On the tab page  Gazetteer
select the database containing the gazetteer data and the project for
which the entries should be synchronized. There are 2 parts for the
synchronization: The Place and the Coordinates. Select one of
these parts for synchronization. To start the query click on the Check
for differences
Gazetteer
select the database containing the gazetteer data and the project for
which the entries should be synchronized. There are 2 parts for the
synchronization: The Place and the Coordinates. Select one of
these parts for synchronization. To start the query click on the Check
for differences  button.
button.
The form will list all detected differences. To update the database
click on the Start update  button.
button.
Maintenance - Gazetteer: Adding links
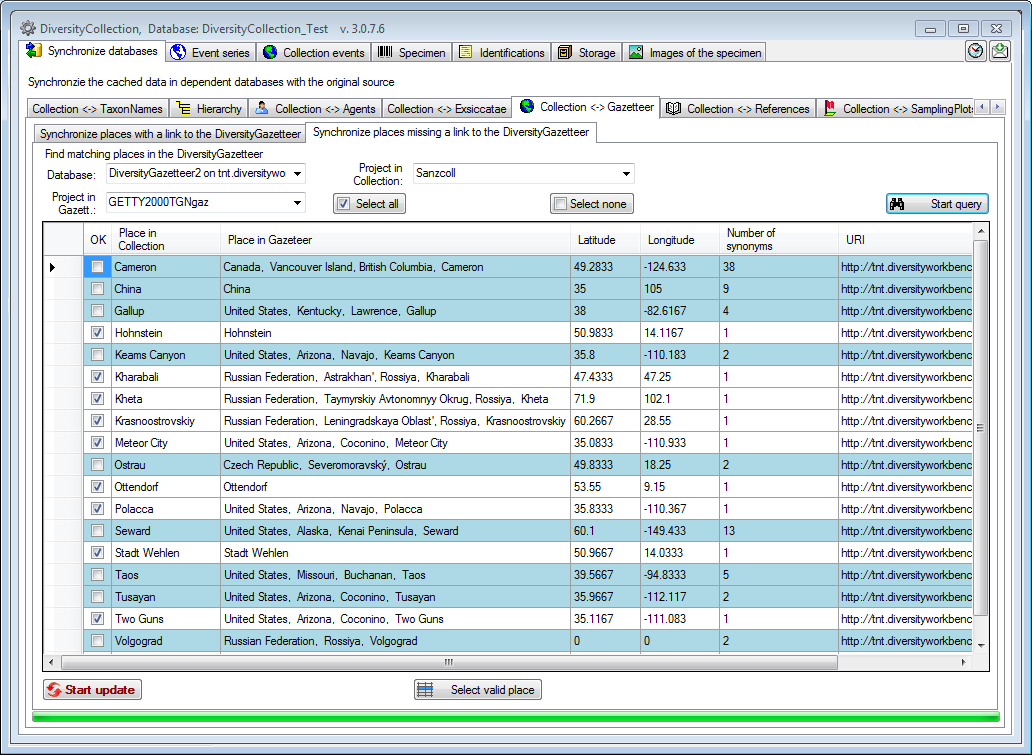
To set links to the DiversityGazetteer for places which are not linked
so far choose the second tab under Collection <→ Gazetteer for
synchronization of places missing a connection to the
DiversityGazetteer. Choose the database and the project and click on the
 Start query button. The results will be listed
as shown below.
Start query button. The results will be listed
as shown below.
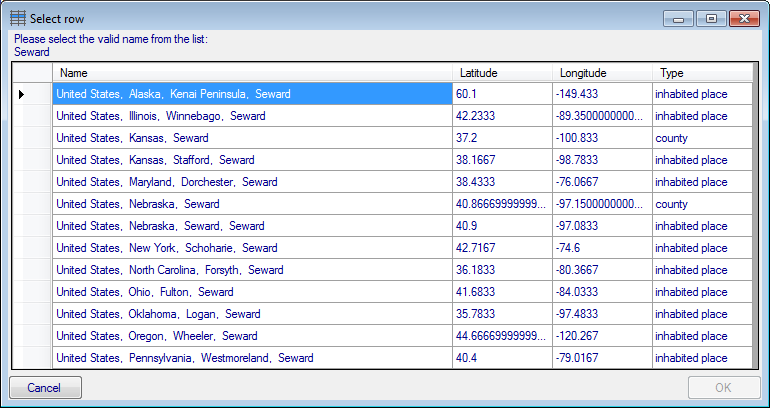
Many places have identical names in different regions. Names where
several results were found are marked in blue. To choose the correct
name among the list of results, click on the  Select valid place button and select the correct name from the list
as shown below.
Select valid place button and select the correct name from the list
as shown below.
Finally, click on the  Start update button to
enter the results in the database.
Start update button to
enter the results in the database.
Maintenance - Identification
Taxonomic names
Taxonomic names may be linked to an external source. To synchronize the
entries for the taxonomic names derived from the module
DiversityTaxonNames choose Administration → Maintenance… from
the menu. A window will open as shown below. Choose the tab
TaxonNames. There are two ways to synchronize taxonomic names. You
may either Synchronize taxonomic names based on the link via an URI
for entries where the link to a taxonomic database is already
established or you may Synchronize taxonomic names missing a
connection where no link is established and you can query for
identical names in one of the databases.
Synchronize taxonomic names based on the link via an URI
The taxonomic names are stored in the field TaxonomicName in the table
Identification. Select a
taxonomic database and a project and taxonomic group which should be
synchronized. Start the query with a click on the Check for
differences  button.
button.
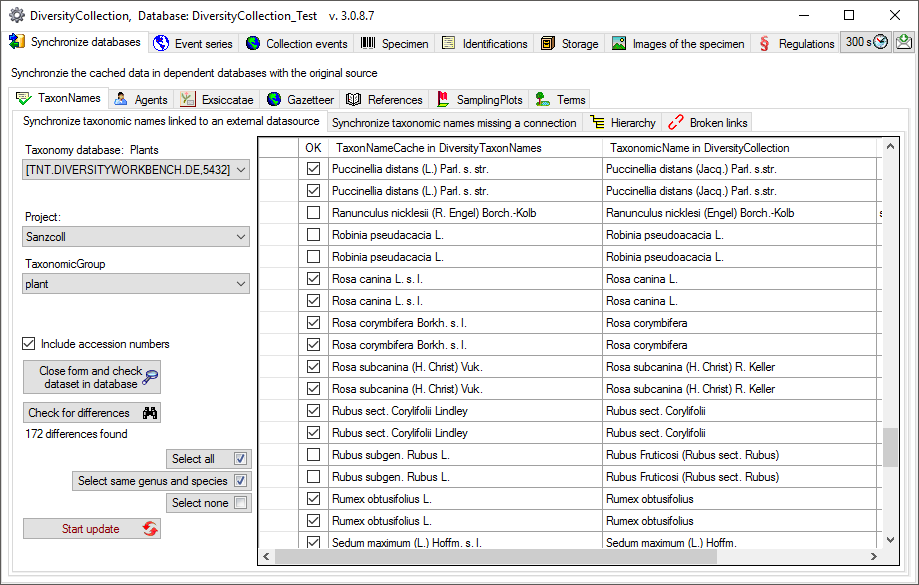
If the taxonomic information has been imported into the
 cache database, you
may use the data from there to speed up the synchronization.
cache database, you
may use the data from there to speed up the synchronization.
The form will list all differences found. With the Select same genus and species  button you can restrict the
selected entries to those where the genus and species are identical in
the source and the entry in collection. The other buttons will either
select all
button you can restrict the
selected entries to those where the genus and species are identical in
the source and the entry in collection. The other buttons will either
select all  or no entry
or no entry  .
.
To update the database click on the Start
update  button. If you wish to have a more
detailed look on your data, you need to check the Include accession number checkbox before starting the query. A button Close form and check dataset in database
button. If you wish to have a more
detailed look on your data, you need to check the Include accession number checkbox before starting the query. A button Close form and check dataset in database  will appear which will
take you back to a single data set in the database.
will appear which will
take you back to a single data set in the database.
Synchronize taxonomic names missing a connection
Select a taxonomic database and a project within this database, a
project in the collection database and a taxonomic group to search for
identical names. Start the query with a click on the Check for identical names button. You have several options to search for fitting
names:
Compare whole name
Search for an exact match. With the option  Excl. auth. you can extend the search for names where the authors are
missing in the identifications.
Excl. auth. you can extend the search for names where the authors are
missing in the identifications.
Compare first parts
Search for names fitting in first parts of the name as shown below. If
the names do not match exactly, you may either update the name (
 Update similar name) in the
collection database or import the name from the DiversityTaxonNames as a
new identification (
Update similar name) in the
collection database or import the name from the DiversityTaxonNames as a
new identification ( Insert as new
name** - see options in the window depicted below).
Insert as new
name** - see options in the window depicted below).
If you choose the option  Restrict to last
identification, the former identifications of an organism will be
ignored.
Restrict to last
identification, the former identifications of an organism will be
ignored.
If you choose the option  Include fuzzy search, the program will try to find names with up to 3 differences.
The search with this option is rather time consuming and should only be
used for a small number of datasets.
Include fuzzy search, the program will try to find names with up to 3 differences.
The search with this option is rather time consuming and should only be
used for a small number of datasets.
In the result list only matching names will be checked (column OK). The
other results will be marked indicating the difference.
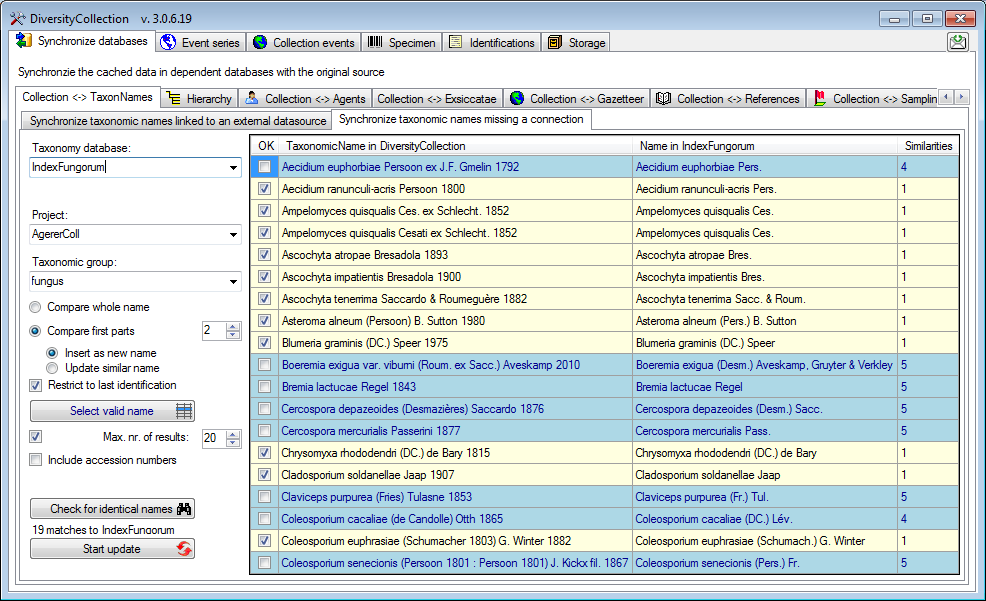
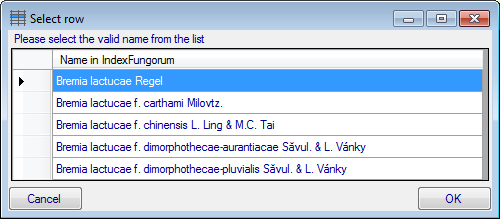
If several similar names were found, these lines will be
marked with a blue background. To select one of the similar names click
on the Select valid name  button. A window
will open as shown below listing all similar names found in Index
Fungorum. Select one name and click OK to use it.
button. A window
will open as shown below listing all similar names found in Index
Fungorum. Select one name and click OK to use it.
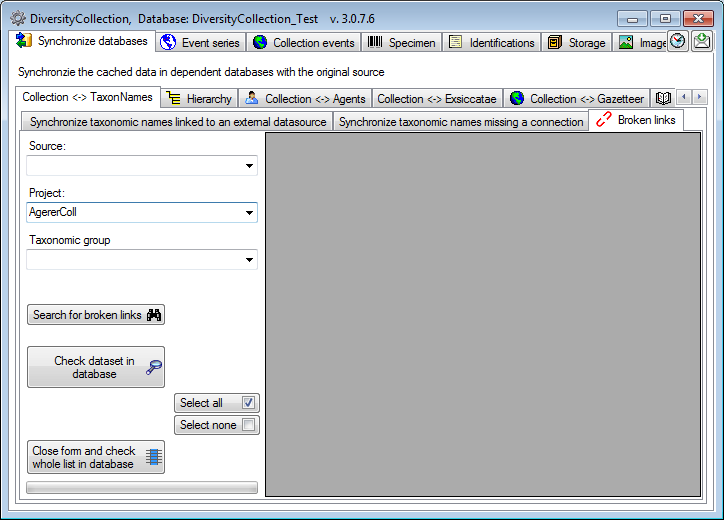
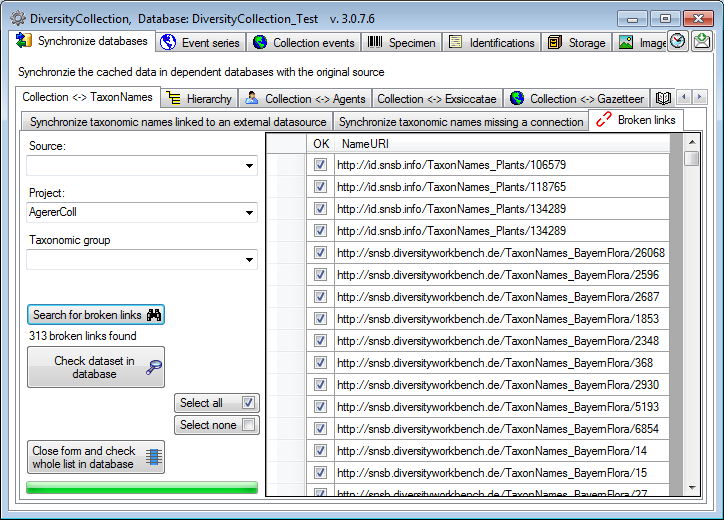
Search for broken links
If for any reason links to webservices or modules are broken  , e.g. when a webservice changes its address, you can search for these (see below).
Please make sure, that you are connected with all
source databases used for the taxa. Otherwise connections to these
sources would be assumed as broken. Choose the Project, for which
you want to search for broken links and optional restrict the search on
a Source and a Taxonomic group. Click on the
, e.g. when a webservice changes its address, you can search for these (see below).
Please make sure, that you are connected with all
source databases used for the taxa. Otherwise connections to these
sources would be assumed as broken. Choose the Project, for which
you want to search for broken links and optional restrict the search on
a Source and a Taxonomic group. Click on the
 button to start the search.
button to start the search.
The broken links will be listed as shown below. With the Check dataset
in database  you can
inspect a single dataset. With the Select all
you can
inspect a single dataset. With the Select all
 and Select none
and Select none
 buttons you can select
resp. deselect all datasets and show the selected datasets with the
Close form and check whole list in the database
buttons you can select
resp. deselect all datasets and show the selected datasets with the
Close form and check whole list in the database
 button in the main
window.
button in the main
window.
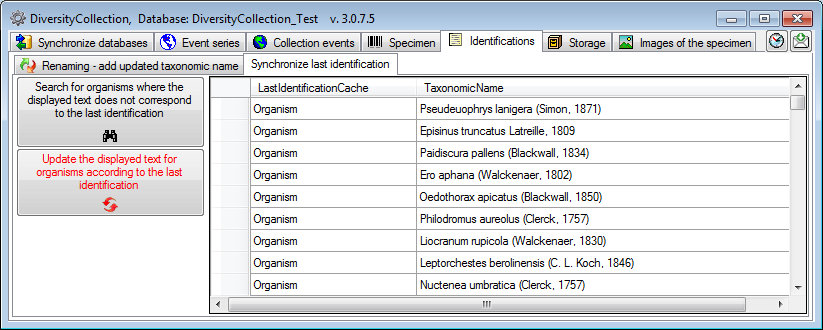
Update the display text for organisms according to last identification
If for any reason the displayed text for an organism does not correspond
to the last identification, this can be fixed under the Synchronize
last identification tab. Just click on the Search
for organisms where the displayed text does not correspond to the last
identification  button to list the deviating display text and then on the Update the displayed text for
organisms according to the last identification
button to list the deviating display text and then on the Update the displayed text for
organisms according to the last identification
 button to synchronize the displayed text with the
last identification.
button to synchronize the displayed text with the
last identification.
Subsections of Identification
Maintenance
Family and order
To synchronize the entries for the family, order and hierarchy of taxa
derived from the module DiversityTaxonNames choose Administration →
Maintenance from the menu. A window will open as shown below. To
synchronize the higher taxonomic entries for entries linked to the
module DiversityTaxonNames choose the tab  Hierarchy. These entries are stored in the fields FamilyCache,OrderCache and HierarchyCache in the table
IdentificationUnit.
Hierarchy. These entries are stored in the fields FamilyCache,OrderCache and HierarchyCache in the table
IdentificationUnit.
Select a taxonomic database, a project and a taxonomic group and then
choose whether you wish to synchronize the family, order or hierarchy.
If you wish to inspect single data sets you need to check the Include
accession number checkbox. Then click on the Check for differences  button to start the query.
button to start the query.
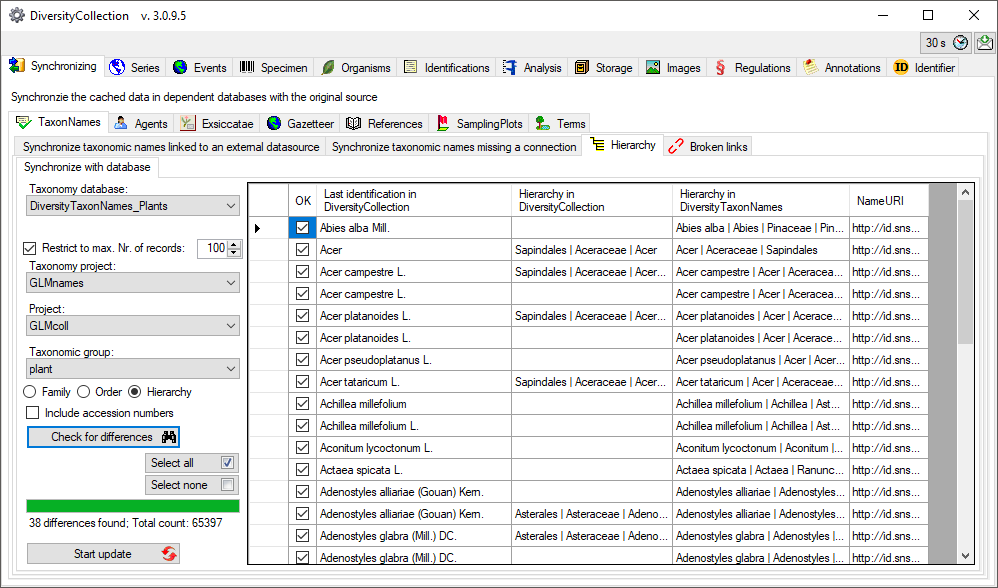
The form will list all differences found as shown above. To update the
database click on the Start update
 button.
button.
Maintenance
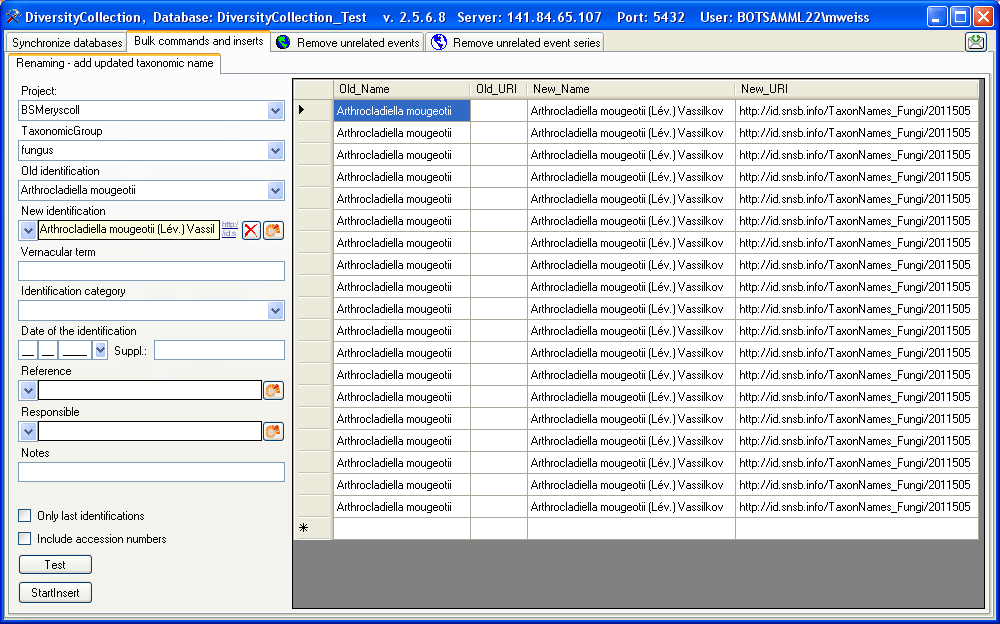
Renaming identifications
If you wish to add updated taxonomic names for specimen in the database,
you can use the bulk insert function. Choose the project, the taxonomic
group and the taxonomic name for which the new identification should be
added. As shown in the window below you may enter additional information
about this identification. By default the Only last identification option is checked to prevent you from searching in
older identifications. If you uncheck this option, the program will
search for any identification, irrespective whether it is the last or an
older identification.
If you wish to check single data sets check the Include accession numbers option.
A new button will appear

which will enable you to close the window and turn to a specimen
selected in the table.
To search for names click on the Test button. The found names
will be listed in the table as shown below. To insert the new
identification click the Start insert button.
Maintenance
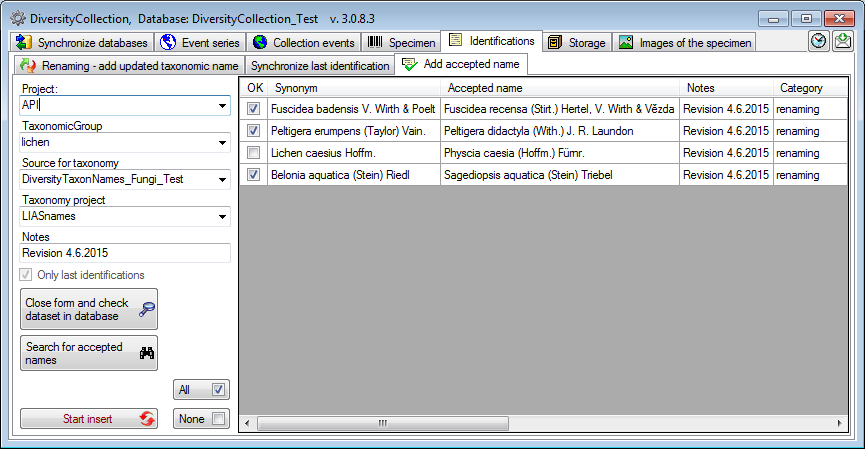
Accepted Names
If you wish to add  accepted taxonomic
names for specimen in the database were the synonyms are used, you can
use the insert function for accepted names. Choose the project, the
taxonomic group for which the new identification should be added as well
as the source for the taxonomic names and the projects within the
taxonomic source from which the new identifications should be taken. As
shown in the window below you may enter additional information in the
notes field.
accepted taxonomic
names for specimen in the database were the synonyms are used, you can
use the insert function for accepted names. Choose the project, the
taxonomic group for which the new identification should be added as well
as the source for the taxonomic names and the projects within the
taxonomic source from which the new identifications should be taken. As
shown in the window below you may enter additional information in the
notes field.
Click on the Search for accepted names  button
to find the accepted names that may be inserted. If you wish to check
single data set, select this dataset in the grid and click on the Close
form and check dataset in database
button
to find the accepted names that may be inserted. If you wish to check
single data set, select this dataset in the grid and click on the Close
form and check dataset in database  button.
button.
If some of the names should not be added, deselect them as shown for the
3. name in the example below. To select or deselect all names you can
use the All  resp. None
resp. None
 buttons. To insert the selected names, click on
the Start insert
buttons. To insert the selected names, click on
the Start insert
 button.
button.
Maintenance
Identification Terms
Vernacular terms may be linked to an external source. To synchronize the
entries for the terms derived from the module DiversityScientificTerms
choose Administration → Maintenance… from the menu. A window
will open as shown below. In the Synchronize databases tab choose
the tab Identification terms. There are two ways to synchronize
scientific terms. You may either Synchronize terms based on the link
via an URI for entries where the link to a scientific terms database
is already established or you may Synchronize terms missing a
connection where no link is established and you can query for
identical terms in one of the databases.
Synchronize terms based on the link via an URI
The terms are stored in the field VernacularTerm in the table
Identification. Select a
database containing scientific terms, a project and the group which
should be synchronized. Finally you have to decide which part should be
synchronized, either the term or the hierarchy where the hierarchy is
available in two variants (upwards or downwards). Start the query with a
click on the Check for differences  button.
button.

The form will list all differences found. The other buttons will either
select all  or no entry
or no entry  .
.
To update the database click on the Start
update  button. If you wish to have a more
detailed look on your data, you need to check the Include accession
number checkbox before starting the query. A button Close form and
check dataset in database
button. If you wish to have a more
detailed look on your data, you need to check the Include accession
number checkbox before starting the query. A button Close form and
check dataset in database  will appear which will
take you back to a single data set in the database.
will appear which will
take you back to a single data set in the database.
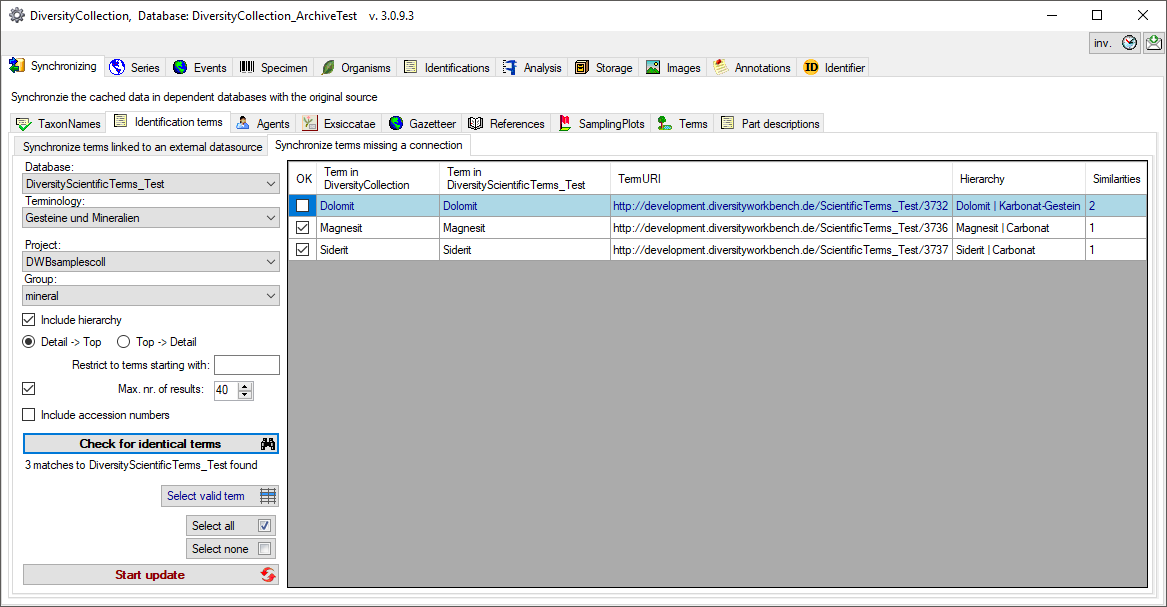
Synchronize terms missing a connection
Select a database containing scientific terms from the list and a
terminology within this database, a project in the collection database
and a group to search for identical terms. For the hierarchy you can
choose between the hierarchy from the term upwards to superior terms
resp. the hierarchy from the top term to the selected term. Start the
query with a click on the Check for identical terms button.
In the result list only terms with a single match will be checked
(column OK).
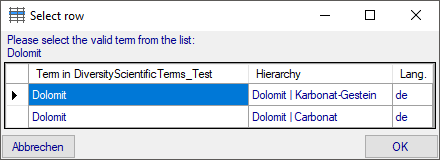
If several similar terms were found, these lines will be
marked with a blue background. To select one of the similar terms click
on the Select valid term  button. A window
will open as shown below listing all similar terms found. Select one
term and click OK to use it.
button. A window
will open as shown below listing all similar terms found. Select one
term and click OK to use it.
Maintenance
Scientific Terms
To synchronize the entries with the module ScientificTerms choose
Administration →  Database →
Database →
 Maintenance from the menu. A window will
open as shown below. On the tab page
Maintenance from the menu. A window will
open as shown below. On the tab page  Synchonizing, you can synchronize the content in DiversityCollection
with the linked resources or search for new links via text.
Synchonizing, you can synchronize the content in DiversityCollection
with the linked resources or search for new links via text.
To synchonize the content for linked data
select the database, the project, the table and the target for
which the entries should be synchronized. There are 2 targets for the
synchronization: The term itself and the hierarchy. There are 3 tables
which may contain links to DiversityScientificTerms:
 Vernacular
terms,
Vernacular
terms,
 Site
properties and
Site
properties and
 Part
descriptions).
Part
descriptions).
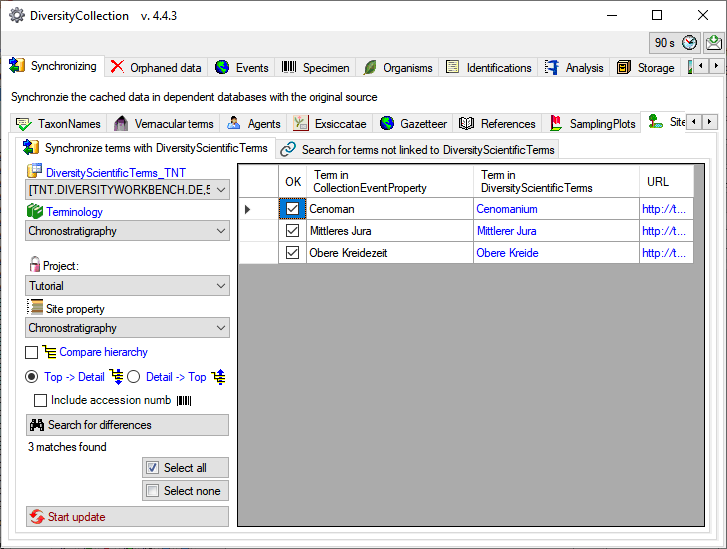
To synchronize  linked data start the query
click on the
linked data start the query
click on the  Search for differences button.
Terms with differing content will be listed as shown below.
Search for differences button.
Terms with differing content will be listed as shown below.
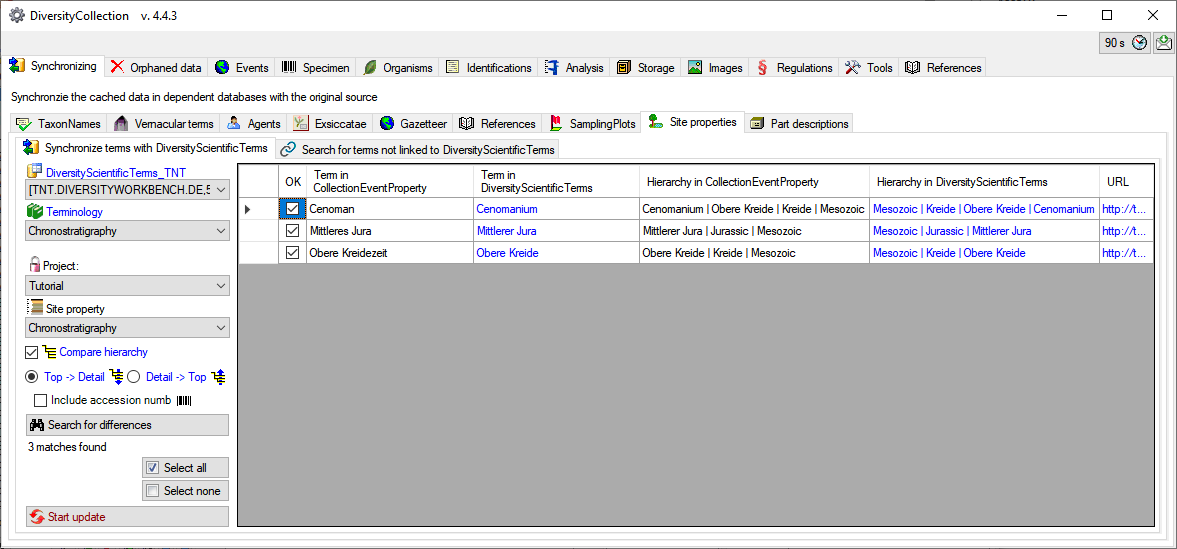
To include the hierarchy in the comparision, check the

 Compare hierarchy
option as well if the hierarchy derived from DiversityScientificTerms should be from
the option
Compare hierarchy
option as well if the hierarchy derived from DiversityScientificTerms should be from
the option  Top→ Detail
Top→ Detail
 or
or  Detail → Top
Detail → Top
 . As shown below 2 more columns will be
included, containing the hierarchies in DiversityCollection and DiversityScientificTerms.
. As shown below 2 more columns will be
included, containing the hierarchies in DiversityCollection and DiversityScientificTerms.
To establish new connections  for terms
corresponding to terms in DiversityScientificTerms (see below)
click on the
for terms
corresponding to terms in DiversityScientificTerms (see below)
click on the  Search for identical terms button.
All matching terms will be listed.
Search for identical terms button.
All matching terms will be listed.
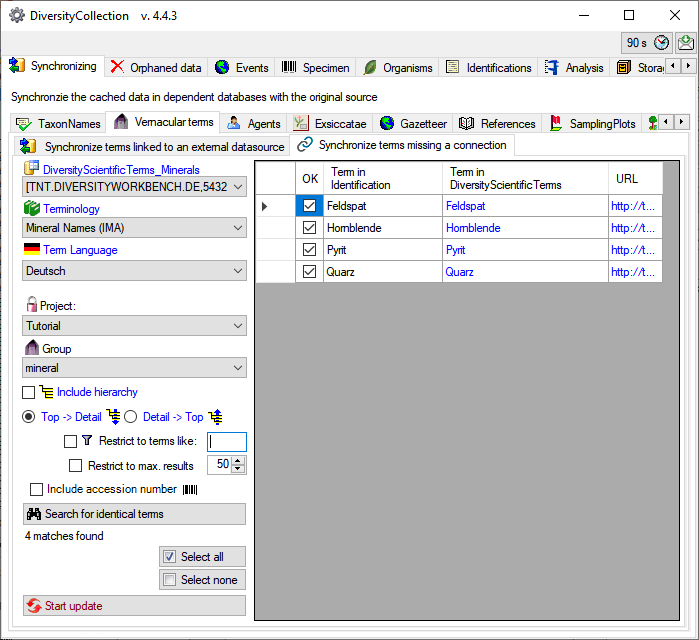
As with the query for linked data, you may include the hierarchy. In
addition there are two options to restrict the search:

 Restrict to terms like
… and
Restrict to terms like
… and  Restrict to max. results … . The
option
Restrict to max. results … . The
option  Include accession number
Include accession number
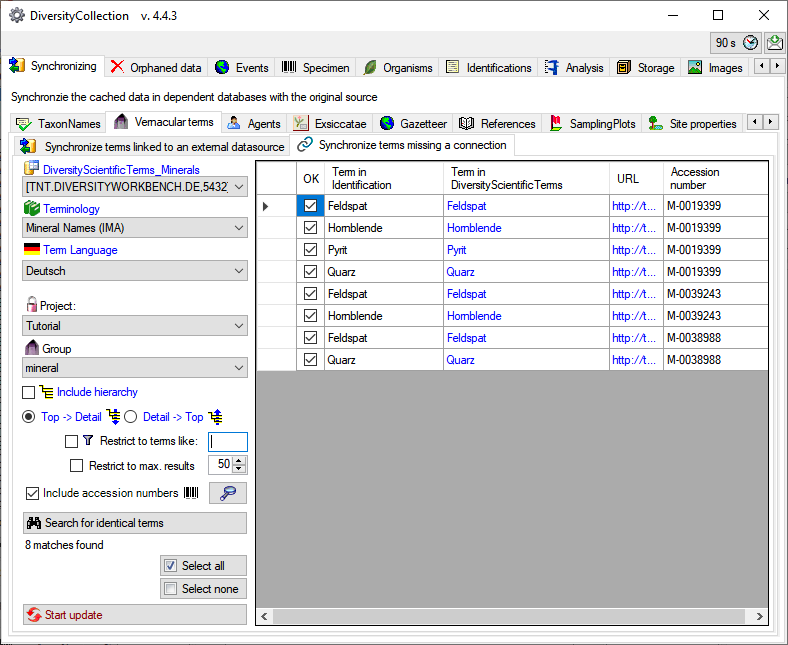
 will split the data according to the specimen
as shown below. With this option set, you can inspect the content of
single specimen in the list using the
will split the data according to the specimen
as shown below. With this option set, you can inspect the content of
single specimen in the list using the  button (see
below).
button (see
below).
In the result list you can select the data that should be included in
the update in the first column (OK  ) by
de/selection of single lines or with the buttons
) by
de/selection of single lines or with the buttons
 Select all resp.
Select all resp.  Select
none. To update the database click on the
Select
none. To update the database click on the  Start update button.
Start update button.
Maintenance
Agents
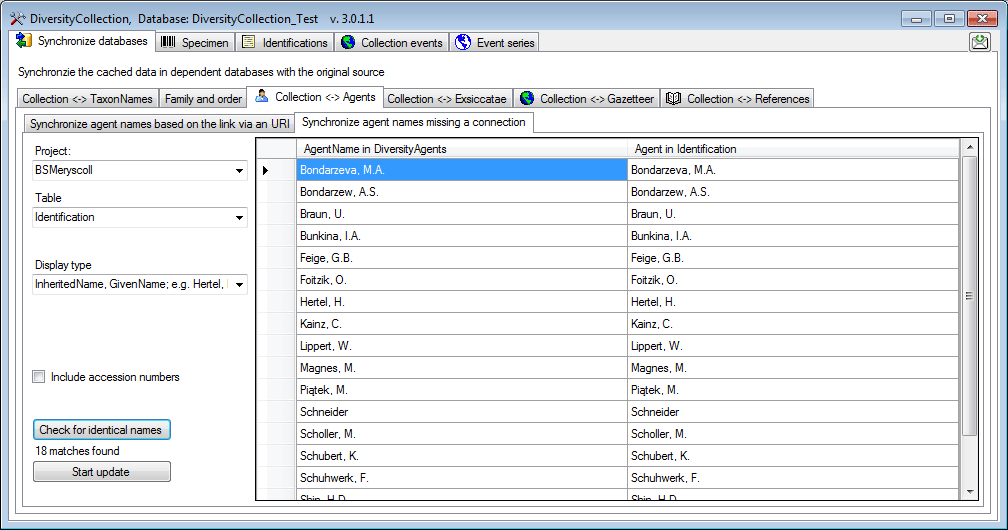
Synchronize via URI
To synchronize the names of collectors, determinators etc. which are
linked to the module DiversityAgents choose Administration -
Maintenance from the menu. A window will open as shown below. On the
tab page Collection <→ Agents select the project for which the
agents should be synchronized. There are 9 tables which may contain
links to DiversityAgents:
Collection,
CollectionAgent,
CollectionEventLocalisation,
CollectionEventProperty,
CollectionSpecimen,
CollectionSpecimenProcessing,
Identification,
IdentificationUnitAnalysis
and Transaction. Choose one of
these tables for the synchronization. Then start the query with a click
on the  button. The type name is written
as defined by the default display type in the module DiversityAgents -
for more information please see the documentation on DiversityAgents.
button. The type name is written
as defined by the default display type in the module DiversityAgents -
for more information please see the documentation on DiversityAgents.
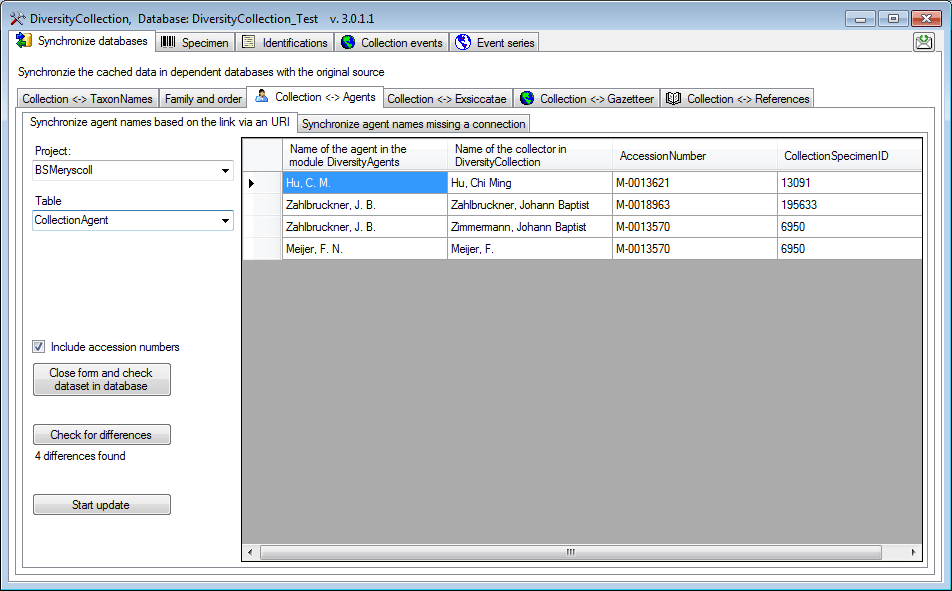
The form will list all differences found. To update the database click
on the  button. If you wish to have a more
detailed look on one of the data sets in the list, you need to check the
“Include accession number” checkbox before starting the query. A button
button. If you wish to have a more
detailed look on one of the data sets in the list, you need to check the
“Include accession number” checkbox before starting the query. A button
 will appear which will take you back to a
single data set in the database.
will appear which will take you back to a
single data set in the database.
Search for possible links
You can either update the cached names for entries which are linked to
DiversityAgents as described above or you can search for possible links
based on the names as shown below. In addition to the options described
above you may choose the display type for the query to find any possible
match as names may be written like “Hertel, H.”, “H. Hertel” etc. 
Next to a search for identical names you can search for similar names.
Enter a search string in the
Like field and click on the  Search for
similar names button. The deteted datasets will be listed as
shown below. For those dataset where there are more than 1 possible link
in the DiversityAgents database you have to choose the correct link with
a click on the Select valid
names button.
Search for
similar names button. The deteted datasets will be listed as
shown below. For those dataset where there are more than 1 possible link
in the DiversityAgents database you have to choose the correct link with
a click on the Select valid
names button.
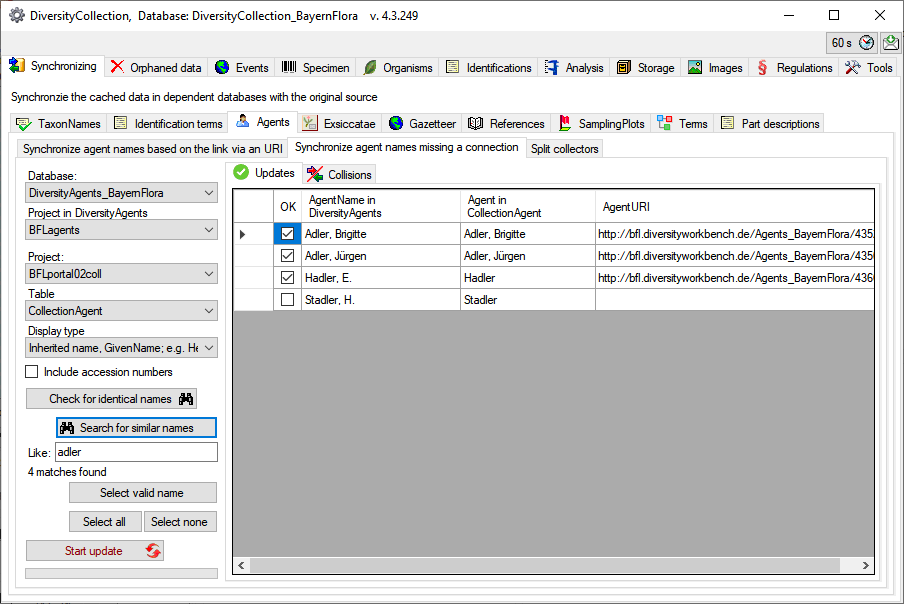
For those dataset where there are more than 1 possible link in the
DiversityAgents database you have to choose the correct link with a
click on the Select valid
names button. A window as shown below will open, where you can
select the valid name. Click on the  button to
show details of an agent.
button to
show details of an agent.
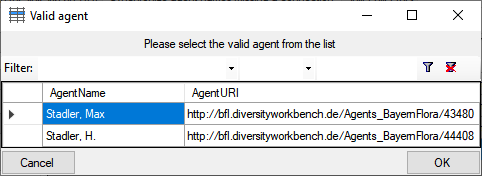
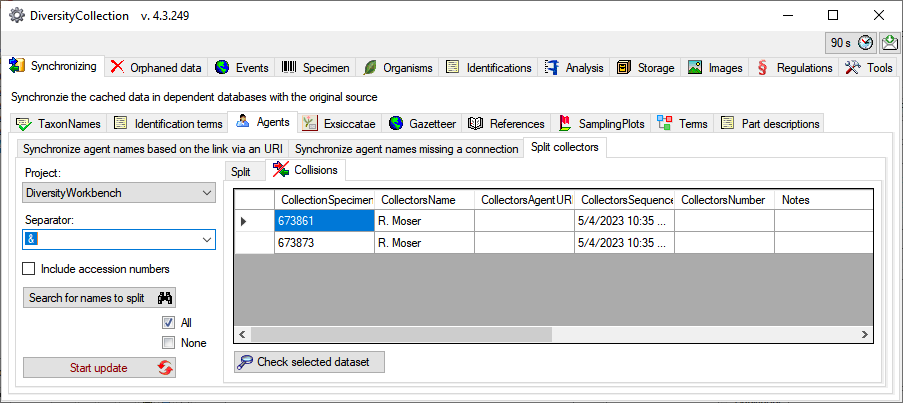
The dataset where the update collides with existing data will be listed
in the tab. Use the
 Check selected
dataset button to inspect these data (see below).
Check selected
dataset button to inspect these data (see below).
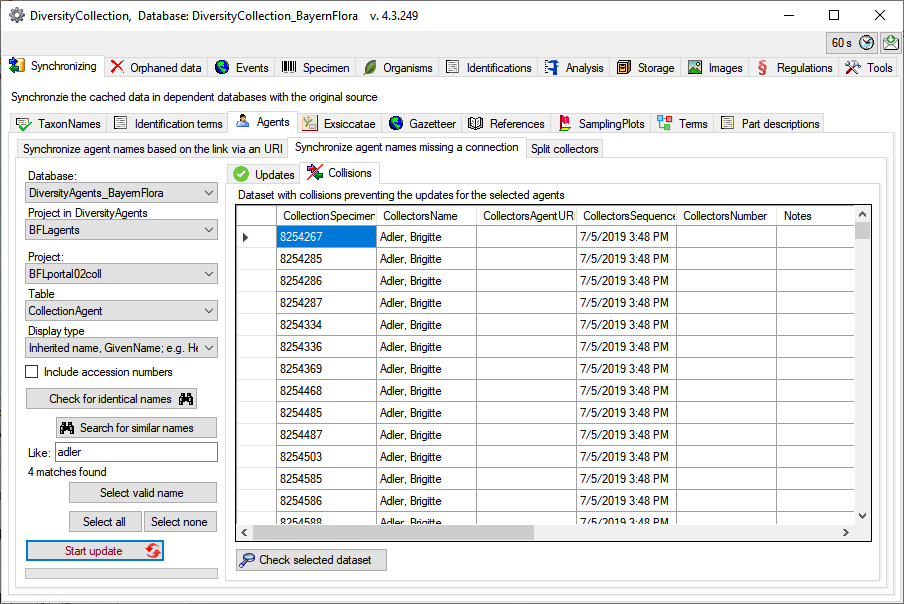
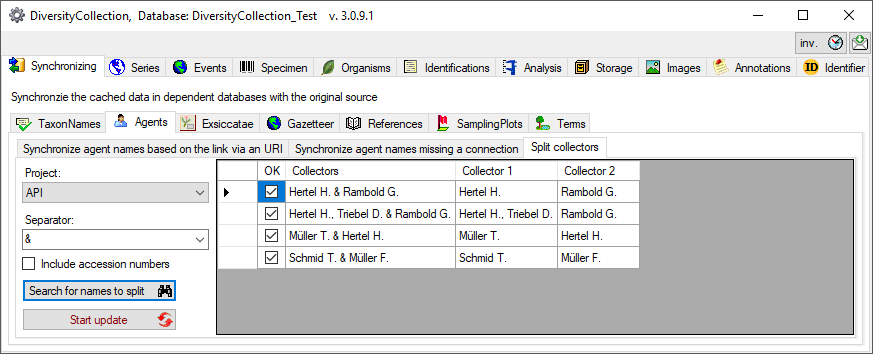
Splitting collectors
If the collectors had been written in one field with separators like &
you can split these to create a separate entry for every collector (see
below). Choose the project, enter the separator or select one of the
predefined separators and click on the  Search for
names to split button. The result will be shown as in the example
below. Click on the
Search for
names to split button. The result will be shown as in the example
below. Click on the  Start
update button to split the collectors. If several separators had
been used you have to do the splitting for every single separator.
Start
update button to split the collectors. If several separators had
been used you have to do the splitting for every single separator.
The dataset where the split collides with existing data will be listed
in the tab. Use the
 Check selected
dataset button to inspect these data.
Check selected
dataset button to inspect these data.
Maintenance
Synchronize
Site properties
Linked to DST
To synchronize the entries derived from the module DiversityScientificTerms
choose → from the menu. On the tab page - - .
Select the  database containing the data, the
database containing the data, the  terminology, the
terminology, the  project and the
project and the  site property for which the entries should be synchronized.
site property for which the entries should be synchronized.
Options:
 Compare hierarchy:
Compare hierarchy:
Click on the button to start the query.
To select or deselect the data that should be updated you can use the buttons
With the button you can synchronize the selected datasets
via Text
To search for data that are not linked to the module DiversityScientificTerms
choose → from the menu. On the tab page - - .
Select the containing the data, the , the  language, the
language, the  project and the
project and the  site property for which the entries should be synchronized.
site property for which the entries should be synchronized.
Options:
 Include hierarchy:
Include hierarchy:
Click on the button to start the query.
To select or deselect the data that should be updated you can use the buttons
With the button you can synchronize the selected datasets
Maintenance
Exsiccatae
To synchronize the abbreviations of the exsiccatal series titles which
are linked to the module DiversityExsiccatae choose Administration →
Maintenance… from the menu. A window will open as shown below. On
the tab page Collection <→ Exsiccatae select the project for
which the titles should be synchronized. The title is stored in the
field ExsiccataAbbreviation in the table
CollectionSpecimen. Select
the project which should be synchronized and start the query by clicking
on the  button.
button.
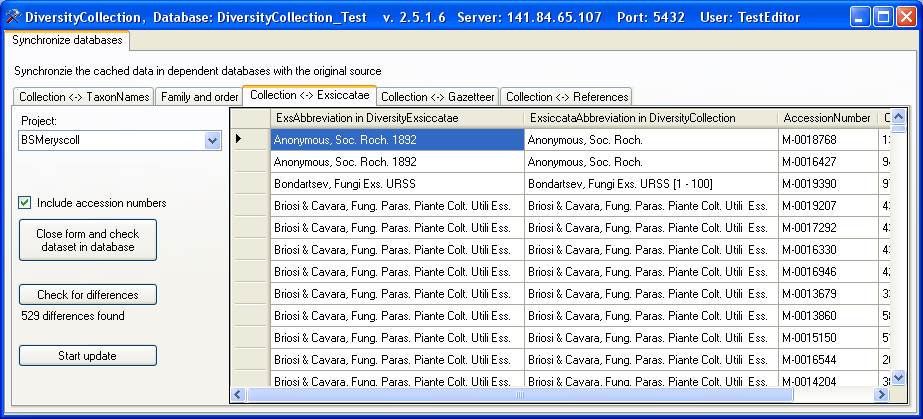
The form will list all differences found. To update the database click
on the  button. If you wish to have a more
detailed look at your data, you need to check the “Include accession
number” checkbox before starting the query. A button
button. If you wish to have a more
detailed look at your data, you need to check the “Include accession
number” checkbox before starting the query. A button
 will appear which will take you back to a
single data set in the database.
will appear which will take you back to a
single data set in the database.
Maintenance
Sampling Plots
To synchronize the entries derived from the module
DiversitySamplingPlots choose Administration → Maintenance from
the menu. A window will open as shown below. On the tab page
Collection → SamplingPlots select the project for which the
entries should be synchronized. There are 2 targets for the
synchronization: The plot identifier (as stored in the field Location1), the Geography and
the cached coordinates in table
CollectionEventLocalisation.
Select the project, one of targets for the synchronization and the
DiversitySamplingPlots database. To start the query click on the
 button.
button.
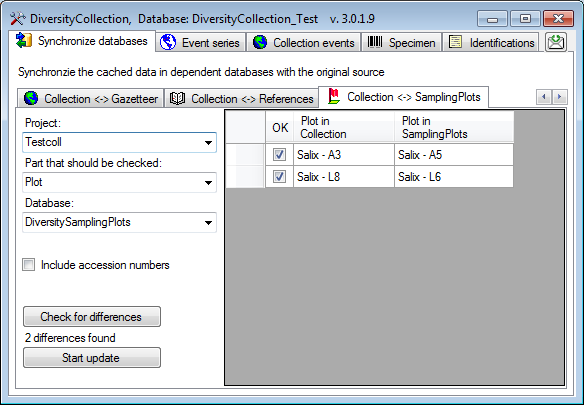
The form will list all differences found. With the first column (OK) you
may deselect certain entries which you do not wish to change. To update
the database click on the  button. If you wish
to have a more detailed look on your data, you need to check the
Include accession number checkbox before starting the query. A
button
button. If you wish
to have a more detailed look on your data, you need to check the
Include accession number checkbox before starting the query. A
button  will appear which will take you back
to a single data set in the database.
will appear which will take you back
to a single data set in the database.
Maintenance
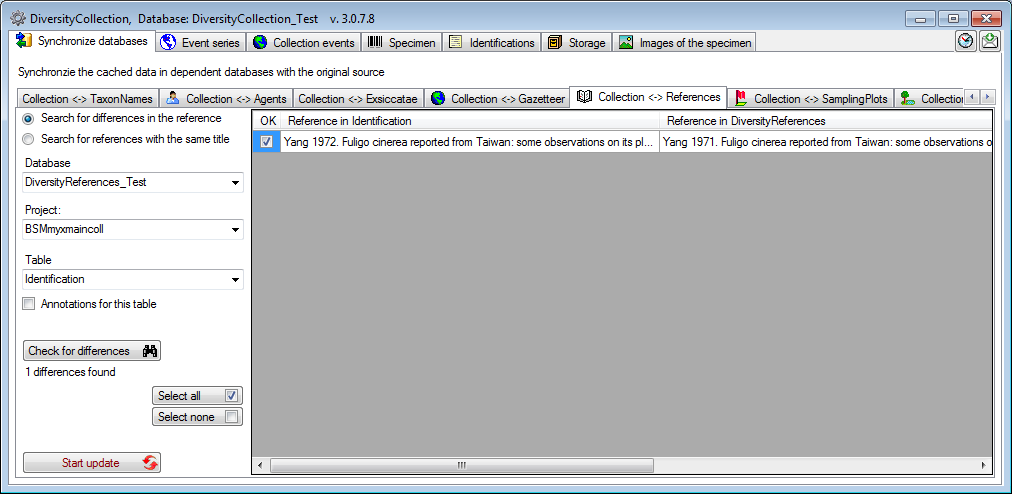
References
To synchronize the titles of the references which are linked to the
module DiversityReferences choose Administration → Maintenance
from the menu. A window will open as shown below. On the tab page
Collection <→ References select the project for which the
reference titles should be synchronized. With the first option you can
 Search for differences in the reference.
There are 3 tables which may contain links to DiversityReferences:
CollectionEvent,
CollectionSpecimen and
Identification. Choose one of
these tables for synchronization. Start the query with a click on the
Check for differences 
The form will list all differences found. To update the data click on
the Start update
 button.
button.
As a second option you can  Search for
references with the same title as found in the module
DiversityReferences (see below).
Search for
references with the same title as found in the module
DiversityReferences (see below).
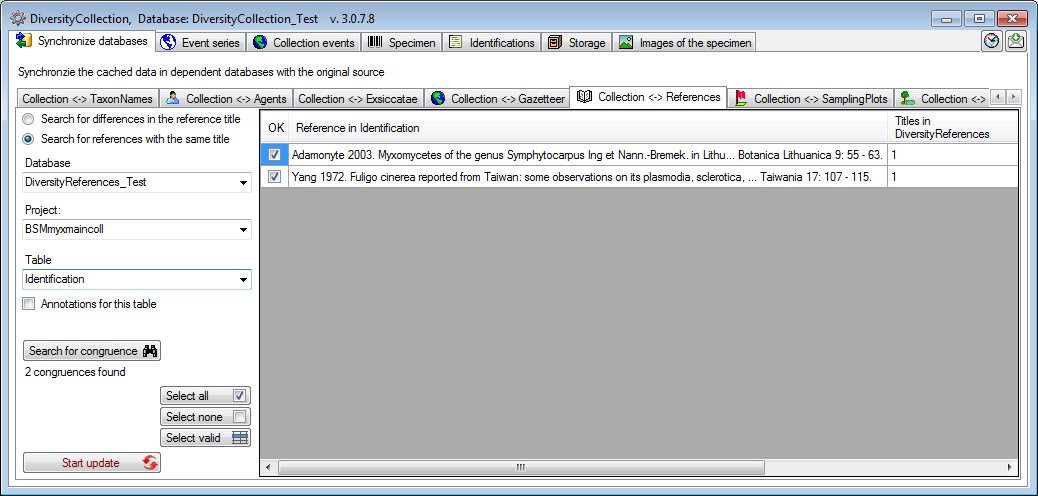
If there is more than 1 reference matching the entry in
DiversityCollection, select the line and click on the Select valid
 button to choose the correct reference.
button to choose the correct reference.
Next to the references in the tables mentioned above, you can perform
the same checks for references stored in the table
Annotation related to the tables
CollectionEvent,
CollectionSpecimen,
CollectionSpecimenPart
and IdentificationUnit. To
use this option, check  Annotations for this
table and select the table that should be checked (see below)
Annotations for this
table and select the table that should be checked (see below)




 button.
button.
 button.
button.
 Select valid place button and select the correct name from the list
as shown below.
Select valid place button and select the correct name from the list
as shown below.



 button you can restrict the
selected entries to those where the genus and species are identical in
the source and the entry in collection. The other buttons will either
select all
button you can restrict the
selected entries to those where the genus and species are identical in
the source and the entry in collection. The other buttons will either
select all  .
. Update similar name) in the
collection database or import the name from the DiversityTaxonNames as a
new identification (
Update similar name) in the
collection database or import the name from the DiversityTaxonNames as a
new identification (

 , e.g. when a webservice changes its address, you can search for these (see below).
Please make sure, that you are
, e.g. when a webservice changes its address, you can search for these (see below).
Please make sure, that you are 
 you can
inspect a single dataset. With the Select all
you can
inspect a single dataset. With the Select all
 button in the main
window.
button in the main
window.










 Database →
Database →
 Maintenance from the menu. A window will
open as shown below. On the tab page
Maintenance from the menu. A window will
open as shown below. On the tab page 


 or
or  . As shown below 2 more columns will be
included, containing the hierarchies in DiversityCollection and
. As shown below 2 more columns will be
included, containing the hierarchies in DiversityCollection and 
 for terms
corresponding to terms in
for terms
corresponding to terms in 
 Restrict to terms like
… and
Restrict to terms like
… and  will split the data according to the specimen
as shown below. With this option set, you can inspect the content of
single specimen in the list using the
will split the data according to the specimen
as shown below. With this option set, you can inspect the content of
single specimen in the list using the 

 button. The type name is written
as defined by the default display type in the module DiversityAgents -
for more information please see the documentation on DiversityAgents.
button. The type name is written
as defined by the default display type in the module DiversityAgents -
for more information please see the documentation on DiversityAgents.
 button. If you wish to have a more
detailed look on one of the data sets in the list, you need to check the
“Include accession number” checkbox before starting the query. A button
button. If you wish to have a more
detailed look on one of the data sets in the list, you need to check the
“Include accession number” checkbox before starting the query. A button







 terminology, the
terminology, the  project and the
project and the 
 language, the
language, the  site property for which the entries should be synchronized.
site property for which the entries should be synchronized.