Edit Description
Editing the description
After starting a database query and selecting an entry in the result
list the dataset is diplayed in the edit window. In the header area the
dataset name, the default project language and the URI of the current
dataset are show (see picture below). Below the header area the
description name may be edited in a text box. Changes of the description
data, e.g. the name, will be visible in the header after saving the
dataset. To check dataset changes, click on the  button to open the history. By clicking button
button to open the history. By clicking button
 you may open a simplified HTML
form to view and edit the description data.
you may open a simplified HTML
form to view and edit the description data.
The main description data may be accessed in the first Mainattributes tab, which is described below. Click on one of the other tabs in the picture below to go directly to the appropriate manual chapter.
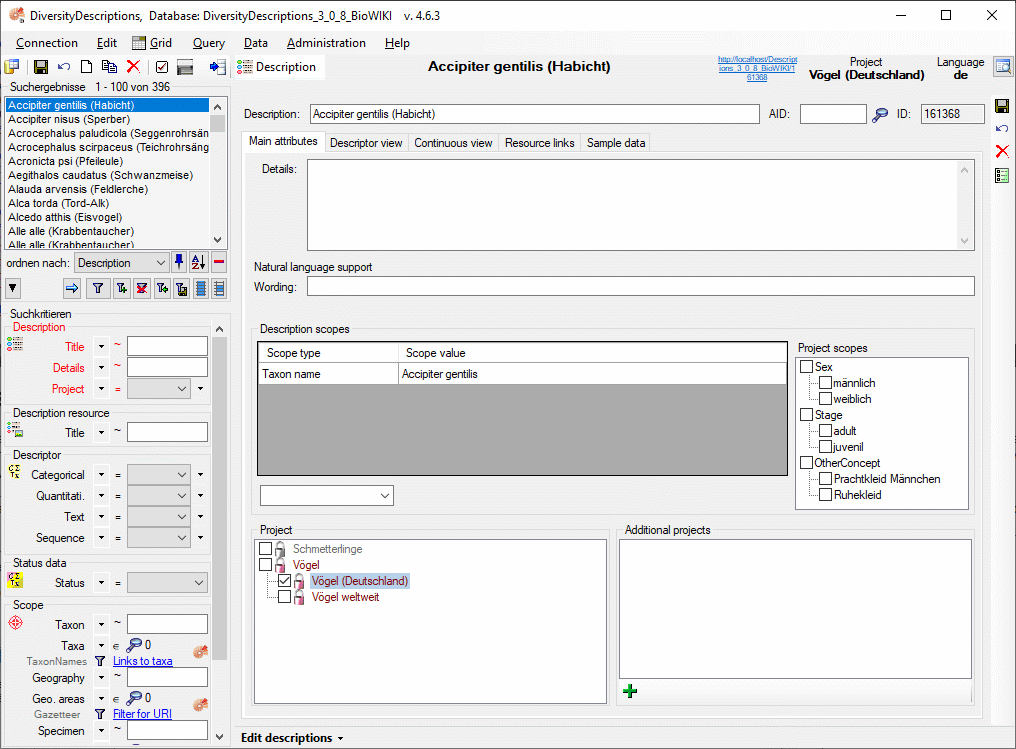
Main attributes tab
The main attributes tab provides in the upper area a field to enter
additional description details and a wording for natural language
support. The optional filed AID may keep an alpha-numeric alternate
ID of the dataset. If used, it should be unambigious within the project,
but this is not checked by the program. Click on  to
get an overview of the used keys.
to
get an overview of the used keys.
Scopes
The next section Description scopes allows specification of
description scopes. This sections consists of the scope table with scope
type and value and a edit section to modify a seleted scope entry or to
enter e new scope. The scope types Geographic ares, Reference,
Specimen, Observation and Taxon name allow input of a
free-form text or reference to the corresponding DiversityWorkbench
module or a web service. The connection to to a module or web service is
done using the button  . After
selecting the entry in the referenced database the text will be taken
over and the backround colour of the scope value field changes to yellow. A further editing of the text will not be
possible (see below).
. After
selecting the entry in the referenced database the text will be taken
over and the backround colour of the scope value field changes to yellow. A further editing of the text will not be
possible (see below).

Additionally the edit section now shows the buttons
 to view the database link and
to view the database link and
 to remove the database link. In the latter case
editing of the scope value will be possible again.
to remove the database link. In the latter case
editing of the scope value will be possible again.
If a scope entry has been selected in the scope table, it can be
modified in the edit section. If you want to enter a new scope value
instead, press the  button at the end of the edit
line. The scope entry in the scope table will be deselected and a
selection box for the scope type appears in the edit section (see
below).
button at the end of the edit
line. The scope entry in the scope table will be deselected and a
selection box for the scope type appears in the edit section (see
below).

You may insert mutiple scope values with links to a DiversityWorkbench
module or a webservice. Therfore enter a new scope value with scope type
Geographic ares, Reference, Specimen, Observation and
Taxon name (or select one, which is not yet linked). Besides the
scope type dropdown box the button  is shown,
which allows insert of multiple scopes (see image below).
is shown,
which allows insert of multiple scopes (see image below).
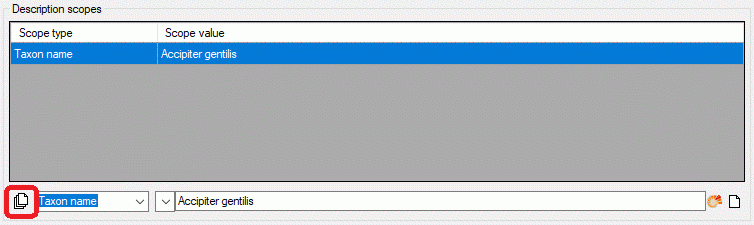
You may insert mutiple scope values with links to a DiversityWorkbench
module or a webservice. Therfore enter a new scope value with scope type
Geographic ares, Reference, Specimen, Observation and
Taxon name (or select one, which is not yet linked). Besides the
scope type dropdown box the button  is shown,
which allows insert of multiple scopes (see image below).
is shown,
which allows insert of multiple scopes (see image below).
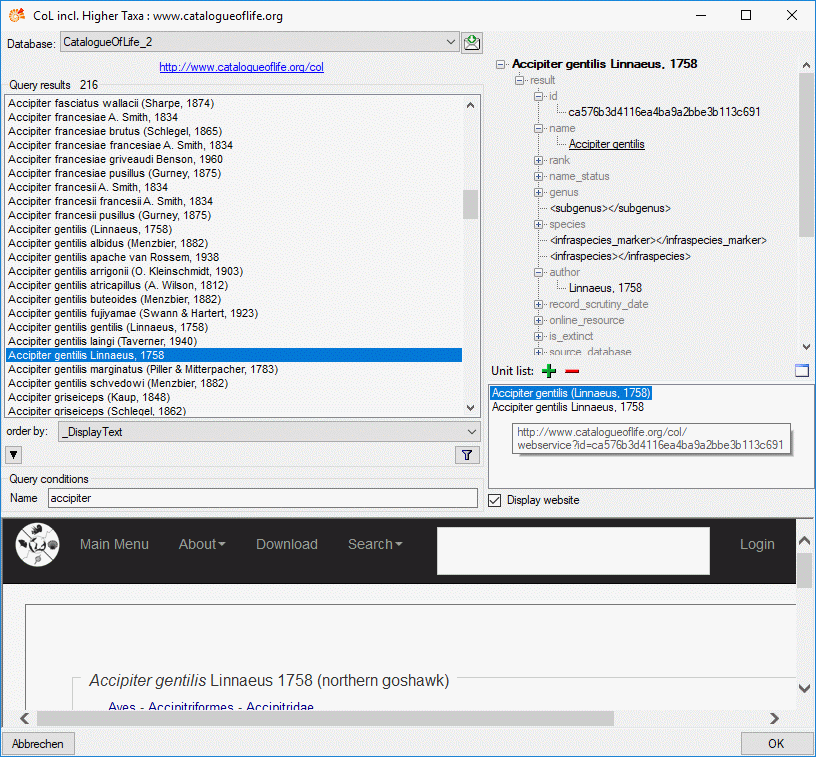
In the remote query panel you find the section Unit list, where you
may add the currently selected query result by clicking the
 button. When you select an entry in the Unit
list, you may remove it by clicking
button. When you select an entry in the Unit
list, you may remove it by clicking  or view the data in a separate window by clicking
or view the data in a separate window by clicking  . When you have collected all query results click the OK button and the scopes
are inserted for the description (see below).
. When you have collected all query results click the OK button and the scopes
are inserted for the description (see below).
Project scopes
Besides the already mentioned scope types that allow input of a free
text or reference to a database entry, the scope types Sex,
Stage, Part and Other scope provide pre-defined scope
values. The values can be administrated as described in the Editing theProject chapter. For each project a different
set of values can be selected as “recommended” values. If for the
current description project scope values are defined/assigned, they are
shown in the right part of the description scopes as Project scopes
(see below).

Selecting or deleting of a scope entry can be easily done by selecting
clickin on the check box before the scope value. You may select rsp.
deselect all scope values of a certain scope type (Sex, State, Part or
Other scope) by clicking the check box of the category. If for a scope
value a detailled description text has been stored, you may view it as
bubble help by moving the mous cursor over the values.
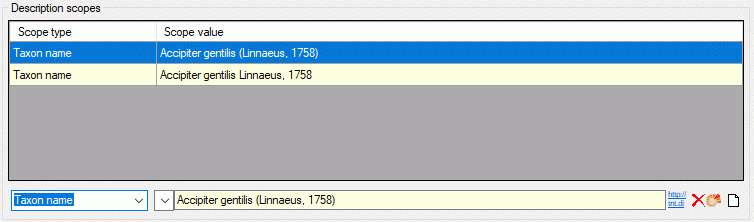
If for a description a scope value is selected, that is not assigned to
its project, the value ist show with yellow
background (see below). Such a situation can occur, if the scope value
has been removed for the project or if a description has been moved to a
different project.

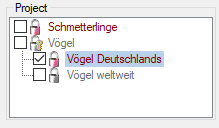
Project
The section Project allows the selection of the project the
description is assigned to. This adjustment has influence of the
available values of scope types Sex, Stage and Other scope, the
available descriptors and recommended modifier/frequency values and
statistical measures of the description. Therefore this unambiguous
adjustment might be seen as the “terminology project” of the description
item. The access rights for a certain user to the projects are
controlled with the login settings (see chapter Loginadministration). When you click on a project
name, you may view some additional project data (see image below
middle).
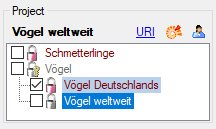
If access to a certain project is restricted to “read only”, it is
displayed with grey colour and symbol
 in the Project section (see pictures
above). A “read only” project cannot be selected for the actual
description. If the selected description has already been assigned to a
“read only” project by another user with appropriate rights, this is
indicated at the right bottom corner of the application window (see
above below right).
in the Project section (see pictures
above). A “read only” project cannot be selected for the actual
description. If the selected description has already been assigned to a
“read only” project by another user with appropriate rights, this is
indicated at the right bottom corner of the application window (see
above below right).
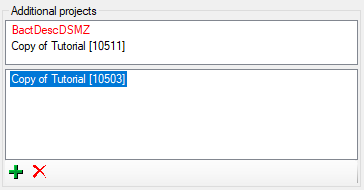
Additional projects
In section Additional projects you may make the description
accessible for additional projects. To assign a description to a project
click on the  button. To remove it from a project,
select the project from the list and click on the
button. To remove it from a project,
select the project from the list and click on the  button. Additional projects without write access are shown in the upper
part of the window, projects with full access in the lower (or only)
part (see image below). If you do not have any access to a project, it
is shown as red text. Be aware that by
entering additional projects you may grant editing rights for single
descriptions to users that have no write access to the current project!
button. Additional projects without write access are shown in the upper
part of the window, projects with full access in the lower (or only)
part (see image below). If you do not have any access to a project, it
is shown as red text. Be aware that by
entering additional projects you may grant editing rights for single
descriptions to users that have no write access to the current project!
Be aware that by entering additional projects you may grant editing
rights for dedicated descriptions to users that have no write access to
the current project! If you have write access to the terminologyproject of a description item and granted access to another
project, where you have read-only access, you nevertheless may withdraw
these rights by removing it from the Additional projects.
Continue with:
Subsections of Edit Description
Edit Description
Continuous view
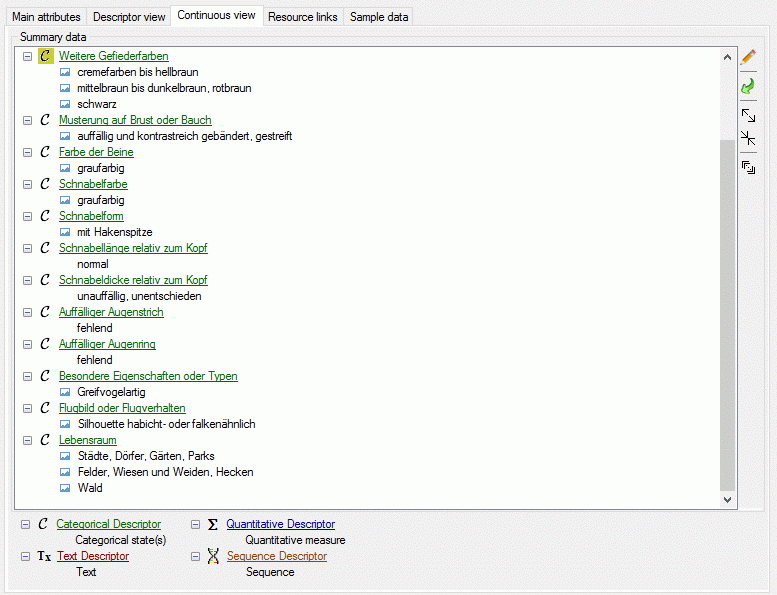
The continuous view tab provides an alternate view on the descriptive
data. In the Summary data only the descriptors are included where
data have been entered. At the right side there is a tool strip. The
displayed buttons will change depending on the selected entry. The
descriptor types 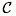 categorical,
categorical, 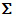 quantitive,
quantitive, 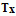 text and
text and  sequence are indicated by different colours
and icons as shown in the example at the bottom. By pressing the
sequence are indicated by different colours
and icons as shown in the example at the bottom. By pressing the
 button on the tool strip this example may be
hidden. Descriptors that are marked as mandatory are displayed with
light red text, descriptors that are
inapplicable are greyed out.
Additionally coloured background indicated possible problems, e.g. if an
inapplicability
rule has been ignored or a not allowed descriptor has been used. A
tool tip text shows the error or warning reason when the mouse curser is
moved over the element.
button on the tool strip this example may be
hidden. Descriptors that are marked as mandatory are displayed with
light red text, descriptors that are
inapplicable are greyed out.
Additionally coloured background indicated possible problems, e.g. if an
inapplicability
rule has been ignored or a not allowed descriptor has been used. A
tool tip text shows the error or warning reason when the mouse curser is
moved over the element.
By pressing the  button the tree view may be
expanded to display the contained categorical states, quantitative
measures or texts. By pressing the
button the tree view may be
expanded to display the contained categorical states, quantitative
measures or texts. By pressing the  button the
tree will be collapsed to descriptor level.
button the
tree will be collapsed to descriptor level.
To insert a descriptor press 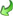 , to delete a
descriptor select the entry and press
, to delete a
descriptor select the entry and press  . If only a
single value shall be removed, select the entry and use the
. If only a
single value shall be removed, select the entry and use the
 button. All these functions can alternatively be
accessed by the context menu by a right-click on the tree node.
button. All these functions can alternatively be
accessed by the context menu by a right-click on the tree node.
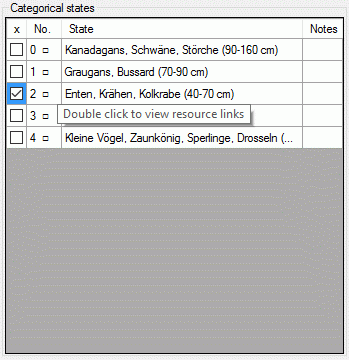
If for a descriptor resources, e.g. pictures, are avaialble, this is
indicated by the background color of the descriptor icon. Categorical
states with resources are marked with icon  (see
first entries in picture above). The resources may be accessed by a
right-click on the tree node and selecting context menu item
(see
first entries in picture above). The resources may be accessed by a
right-click on the tree node and selecting context menu item
 View resources (see picture above).
View resources (see picture above).
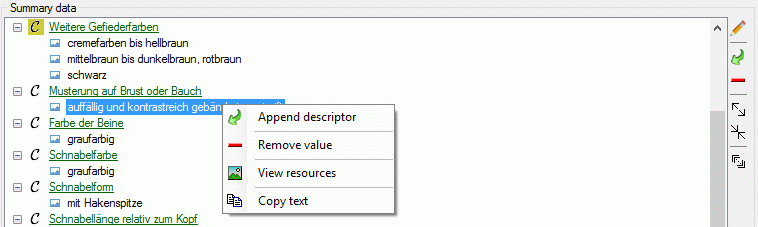
Structured descriptor view
If for the project a structured descriptor tree is defined, i.e. a
descriptor tree that contains at least one descriptor tree node, the
Descriptor tree: drop-down list is shown below the descriptor tree
(see image below left). If a descriptor tree is selected, the descriptor
tree node hierarchy will be included in the output and the descriptors
will be arranged accordingly. The names of the descriptor tree nodes and
the descriptors will be reduced by parts contained in the supriour
hierarchy nodes (see image below right).
If for a descriptor tree node resources are available, this is indicated
by the grey background color of the
descriptor tree icon. The resources may be accessed by a right-click on
the tree node and selecting context menu item  View resources. Additional descriptor tree node information, e.g. an
optional abbreviation or details text can be viewed as bubble help by
placing the mouse cursor over the item.
View resources. Additional descriptor tree node information, e.g. an
optional abbreviation or details text can be viewed as bubble help by
placing the mouse cursor over the item.
Editing summary data
A click on  opens an edit window at the right part
of the tab. This window provides the same functions as available in the
descriptor view tab. The picture below shows the resource linked to a
categorical state that can be opened by doule-clicking the state ID in
the Categorical states section.
opens an edit window at the right part
of the tab. This window provides the same functions as available in the
descriptor view tab. The picture below shows the resource linked to a
categorical state that can be opened by doule-clicking the state ID in
the Categorical states section.
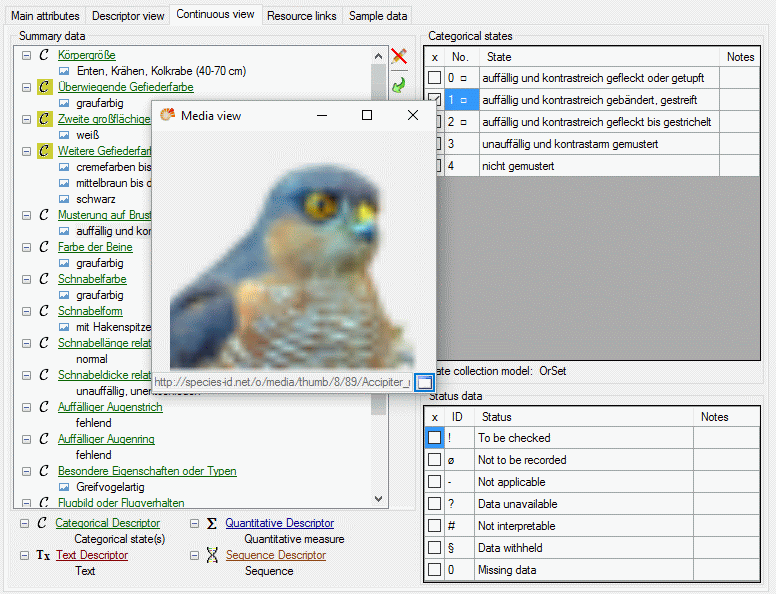
Continue with:
Edit Description
Descriptor view
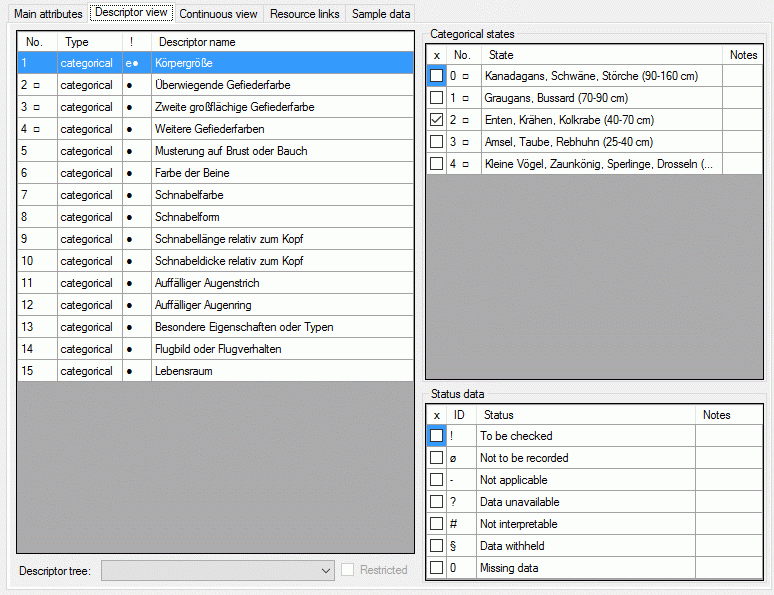
The left part of the descriptor view tab shows all descriptors available
for the description’s project. The entries in column “!” indicate some
additional information about the descriptor:
- Descriptor is referenced in the summary data (●)
- Descriptor is referenced in the status data (○)
- Descriptor is mandatory, values must be specified (!)
- Descriptor is inapplicable because of other specified
descriptors/categorical states (x)
- Descriptor is exclusive, only one categorical states shall be selected
(e)
If resources are linked to a descriptor, the entry is marked by a square
(□) behind its sequency number and a tool tip appears when the mouse
cursor is moved over the “No.” column (see below). By double-clicking
the sequence number a “Media view” window can be opened to view the
resources.
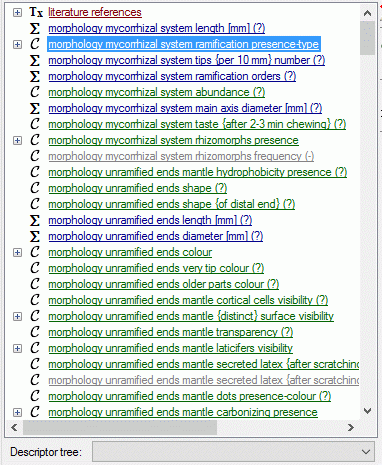
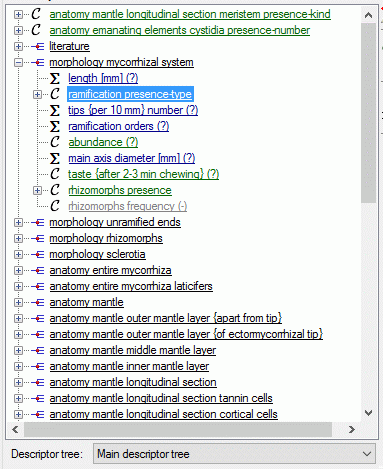
If for the project descriptor trees are defined, the Descriptor
tree: drop-down list and Restricted check box are shown below the
descriptor list (see image below left). If a descriptor tree is
selected, the names of the descriptor tree node hierarchy is included in
the descriptor names (see image below right). This feature is useful, if
several descriptors with ambigious names exist the can now be
distinguished. E.g. two descriptors “colour”, one at descriptor tree
node “Leaf”, the other at descriptor tree node “Blossom” will become
“Leaf colour” and “Blossom colour”. If the descriptor name already
includes the hierarchy, the duplicate parts will be filtered. In the
mentioned example the descriptor names “Leaf colour” and “Blossom
colour” will not be changed, if the descriptor tree is selected.
If additionally the Restricted option is set, only the descriptors
included in the selected descriptor tree are displayed (see image below
right, missing descriptor number 14). So you have the option to create
several descriptor trees that include only special aspects of the
description’s property (e.g. morphology, molecular biology and so on) to
get a clearly arranged descriptor list.
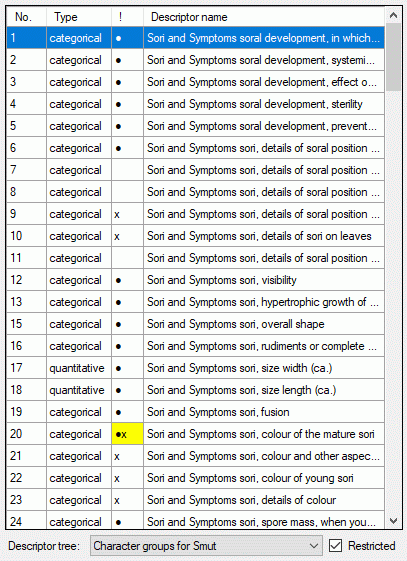
Editing summary data
The right part of the descriptor view tab shows at the bottom the
Status data values of the selected descriptor. It is possible to
select several values for each descriptor. In the “Notes” column
additional information concerning the selected data status may be
entered. By a double click on the “Notes” entry a separate edit window
can be opened.
Depending on the descriptor type you can find at the top either
Categorical states (see large picture above), Statistical
measures, Descriptive text or Molecular sequence (see pictures
below). In the “Notes” columns additional information concerning the
specific categorical state rsp. statistical measure may be entered. By a
double click on the “Notes” entry a separate edit window can be opened.
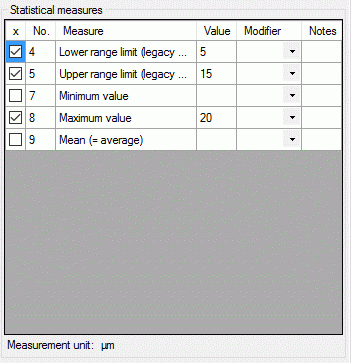
To enter a statistical measure value click in the “Value” field and
enter the value, the check mark in field “x” will be set automatically.
If recommended modifier values are defined (see “Editing the
Descriptor” and “Editing the
Project”), a value may be selected from the drop down
list “Modifier”. If no modifier values are available, the appropriate
table column may be hidden. In the “Notes” field an additional text may
be added. By double-clicking the “Notes” field the Extended texteditor can be opened.
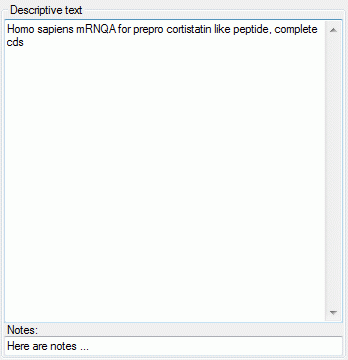
The descriptive text may be edited diretly in the window, which
shows the pure text without any formatting. In the notes field an
additional text may be entered. By double-clicking the text fields the
Extended text editor can be opened.
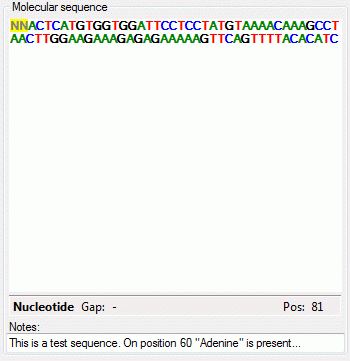
The molecular sequence window displays in the status line below the
edit area the sequence type (“Nucleotide” or “Protein”), the symbol
length (“Len:”) in case of proteine sequences, the gap symbol (“Gap:”)
if defined, the long text of the actual element and the position
(“Pos:”) (see image below). In case of three-letter proteine symbols the
position is the number of the amino acid symbol, not the character
position. By pressing the TAB key the cursor jumps to the next
symbol.

When characters are entered in the edit area that are recognized as
valid symbols, they will automatically
be formatted. One-letter symbols are converted to upper case characters,
three-letter symbols are converted according the “Xxx” schema, e.g.
“Ala” for “Alanine”. The sybols for the nucleotide bases "Adenine", "Cytosine", “Guanine”, "Thymine" and "Uracil" are dispayed with different colors.
Ambiguity symbols and gap symbols are shown in grey color. If the ambiguity symbols have not
been enabled in the descriptor data, they will be displayed with yellow background. Symbols that have not been
recognized are displayed with red
background.
The molecular sequence window offers some additional functions available
in the . It may be
opened by moving the mouse cursor over the control header (“Molecular
sequence”, see window below left) or from the context menu (right-click
on the sequence edit area, see windows below right). With menu item
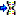 Import you may read
the seqeunce data from a file and insert them to the description. With
menu item
Import you may read
the seqeunce data from a file and insert them to the description. With
menu item  Export
you may export the actual sequence data to a file.
Export
you may export the actual sequence data to a file.
By double-clicking the molecular sequence area or from sequence menu
 Open →
Open →  Sequence a
separate edit window may be opened (see image below). On the top of the
window there is the drop-down menu Insert symbol: where a value may
be selected from a list. After selecting the value press the [ENTER]
key or click on
Sequence a
separate edit window may be opened (see image below). On the top of the
window there is the drop-down menu Insert symbol: where a value may
be selected from a list. After selecting the value press the [ENTER]
key or click on 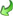 to insert the selected symbol at
the cursor position.
to insert the selected symbol at
the cursor position.
To select a categorical state set a check mark in field “x” with a
mouse click. If recommended modifier or frequency values are defined
(see Editing the Descriptor and Editing the
Project), values may be selected from the drop down
lists “Modifier” rsp. “Frequency”. If no modifier or frequency values
are available, the appropriate table column may be hidden. In the
“Notes” field an additional text may be added. By double-clicking the
“Notes” field the Extended text editor can be
opened.
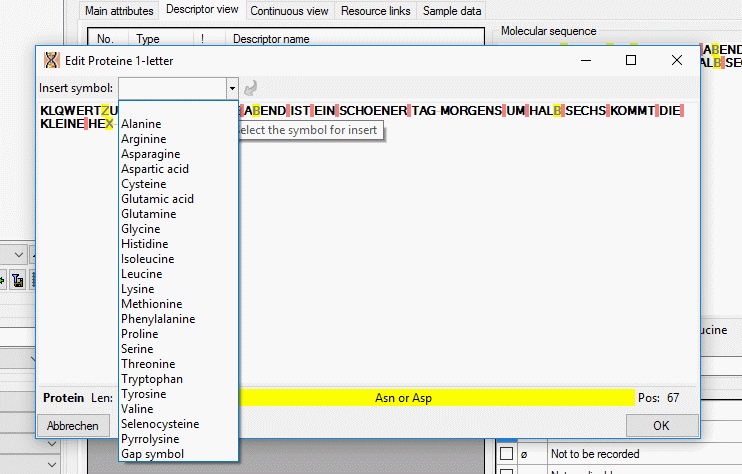
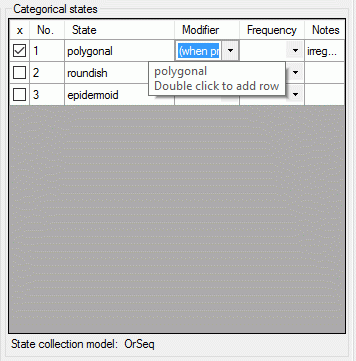
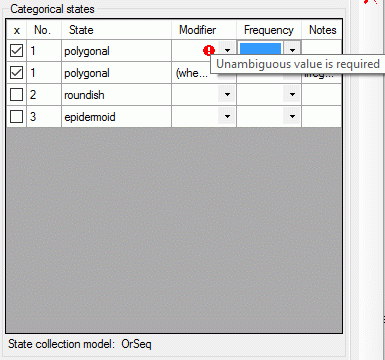
If a “Modifier” or “Frequency” value is selected, the multiple
specification of a dedicated categorical state is possible. A tool tip
offers to add a row by doouble-click, if mouse cursor is moved over the
“State” field (left picture below, picture in the midddle after double
click). Multiple specifications of one categorical state are only
allowed if they have been assigned different modifier and frequency
values. Inputs offending these restrictions will be ignored an a cell
error will indicate the problem (right picture below).
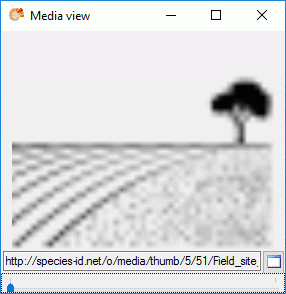
If resources are linked to a categorical state a tool tip appears when
the mouse cursor is moved over the ID (left picture below). By
double-clicking the ID a “Media view” window can be opened to view the
resources (right picture below).
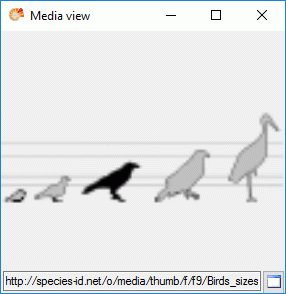
If more than one resources have been assigned to one entry, a slider
will be visible at the bottom of the “Media view” window. To switch
through the available resources the slider can be moved using the mouse
or the right and left arrow keys (see below).
Continue with:
Edit Description
Export sequence
With this form you can export data from the database to a file. Choose
 Export from the sequence
menu to open the window
for the export. Format specifies the file format, currently
FASTA and FASTQ are supported.
Export from the sequence
menu to open the window
for the export. Format specifies the file format, currently
FASTA and FASTQ are supported.
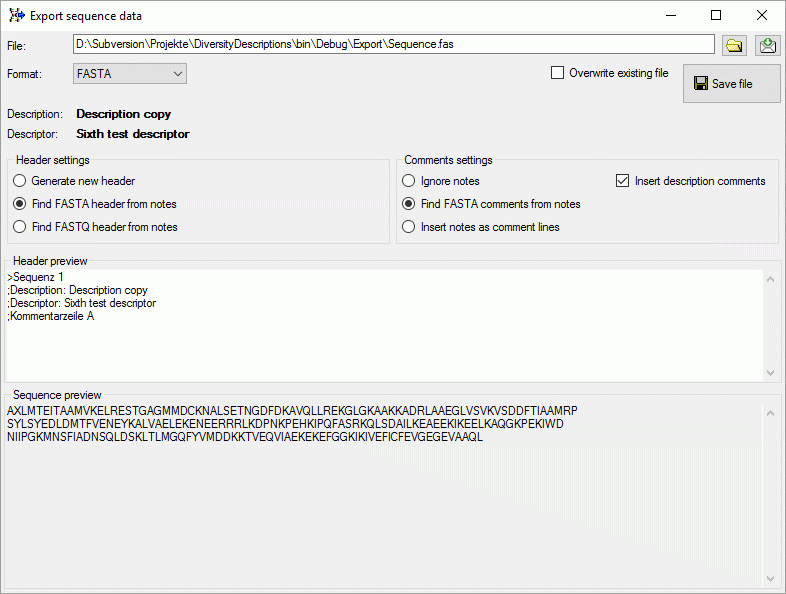
In the Header settings you may specify if a new header shall be
generated or if the notes shall be searched for a FASTA rsp. FASTQ
header. In case of FASTA export you may specify in the Comments
settings section if FASTA comment lines from the notes (starting with
“;”) shall be inserted after the header, the whole notes shall be
inserted as FASTA comments or if the notes shall be ignored. If Insert
description comments is checked, two comment linsed wit the
description and descriptor title will be generated.
In the Header preview section you may check and edit the resulting
header, in the Sequence preview section the converted sequence in
one-letter notation is displayed.
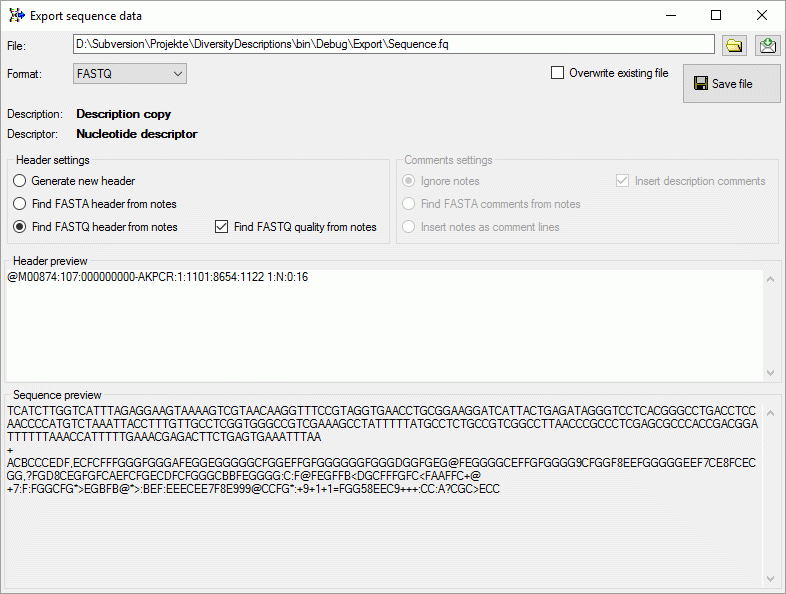
In case of FASTQ export the Comments settings are irrelevant. If the
notes include a QUALITY STRING sections, it is inserted in the FASTQ
output (see image below). If no quality string can be found or the
Find FASTQ quality from notes has been unchecked, the lowest
quaality value “!” is inserted.
Click on the  button to
select the file where the data shall be exported. By default the data
are appended at the end of the selected file, check the Overwrite
existing file option to overwrite it. Finally click the
button to
select the file where the data shall be exported. By default the data
are appended at the end of the selected file, check the Overwrite
existing file option to overwrite it. Finally click the
 Save file button to write the data.
Save file button to write the data.
Continue with:
Edit Description
HTML
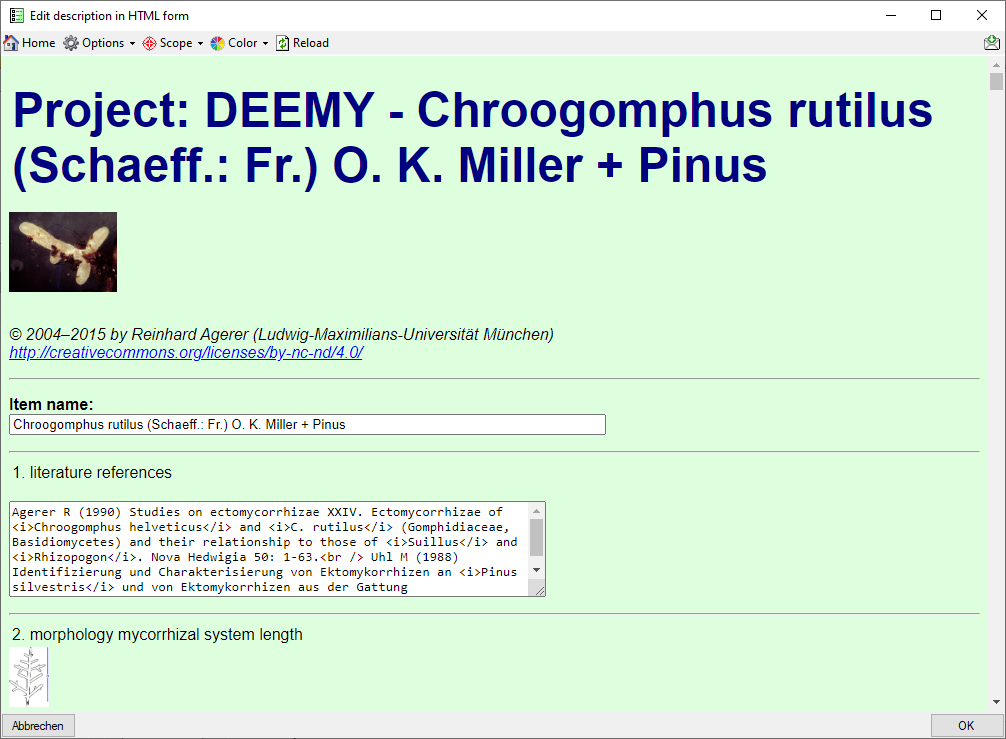
With this form you can easily edit the description data in an HTML form.
After starting a description query and selecting an entry choose Edit
->  Edit as HTML form … from the
menu or click on the
Edit as HTML form … from the
menu or click on the  button at the
right side of the main Diversity Descriptions window. A window as shown
below will be opened.
button at the
right side of the main Diversity Descriptions window. A window as shown
below will be opened.
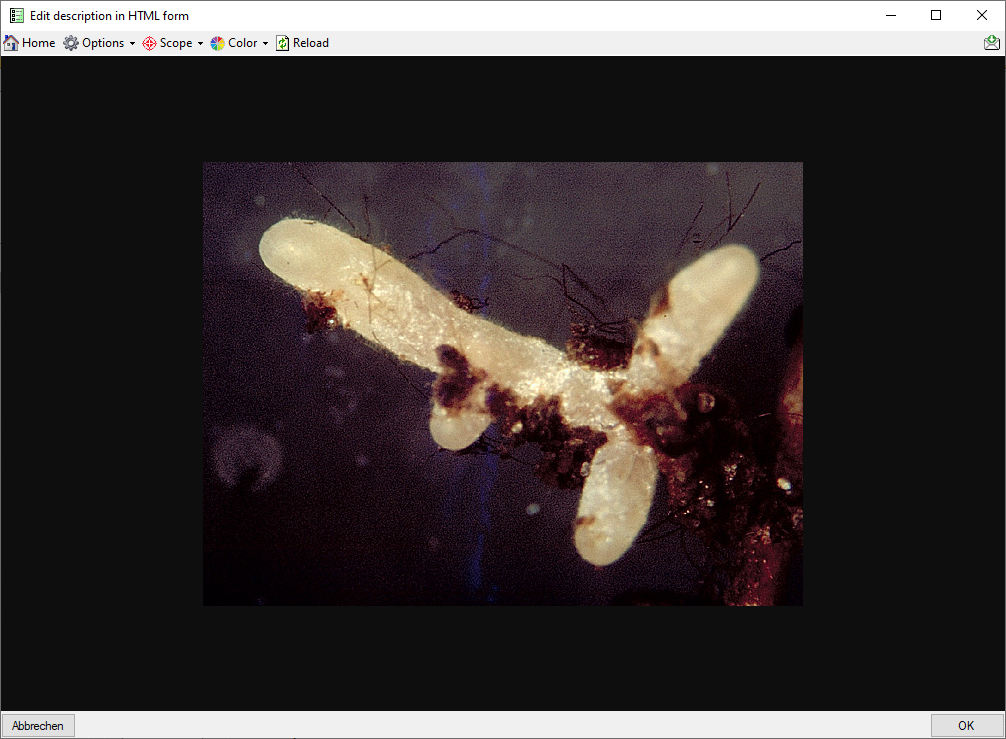
When you move the mouse cursor over embedded
pictures, they are slightly magnified, by clicking on the resource, you
may show it in full size (see below). Use
button  Home to return to the HTML form.
Home to return to the HTML form.
With drop down button  Options you may
insert some additional input fields in the form, e.g. item details or
notes. With drop down button
Options you may
insert some additional input fields in the form, e.g. item details or
notes. With drop down button  Scope you may
insert input fields for the vasiour scope types and drop down button
Scope you may
insert input fields for the vasiour scope types and drop down button
 Color lets you select the background color.
All that changes come into affect, when you reopen the form or redraw
the HTML form using the
Color lets you select the background color.
All that changes come into affect, when you reopen the form or redraw
the HTML form using the  Reload button.
Reload button.
After clicking the  Reload button you will be
asked if you want to discard all changes. By selecting No the HTML
form data will be written into the local data store, otherwise all
inputs will be reset. To exit the form and save all changes in the local
data store, click the OK button. To ignore your changes, click the
Cancel button and you will be asked if you want to discard all
changes. The HTML form utility works completely on the local data store.
I.e. if you edited the description data in the main form of Diversity
Descriptions and start the HTLM form editor without saving the data, all
changes are only present in the application dataset. To store the data
in the database, click the
Reload button you will be
asked if you want to discard all changes. By selecting No the HTML
form data will be written into the local data store, otherwise all
inputs will be reset. To exit the form and save all changes in the local
data store, click the OK button. To ignore your changes, click the
Cancel button and you will be asked if you want to discard all
changes. The HTML form utility works completely on the local data store.
I.e. if you edited the description data in the main form of Diversity
Descriptions and start the HTLM form editor without saving the data, all
changes are only present in the application dataset. To store the data
in the database, click the  save button of the main
form!
save button of the main
form!
Edit Description
Molecular sequence
The editor for sequence data recognizes predefined symbols for
nucleotide and protein sequences according the
IUPAC definitions.
Nucleic acid symbols
|
|
| Symbol |
Name |
| A |
Adenine |
| C |
Cytosine |
| G |
Guanine |
| T |
Thymine |
| U |
Uracile |
| W |
Weak (A or T) |
| S |
Strong (G or C) |
| M |
aMino (A or C) |
| K |
Keto (G or T) |
| R |
puRine (G or A) |
| Y |
pYrimidine (C or T) |
| B |
not A (B comes after A) |
| D |
not C (D comes after C) |
| H |
not G (H comes after G) |
| V |
not T (V comes after T and U) |
| N |
No idea (not a gap) |
The symbols with grey background are
ambiguity symbols. The difference between “N” and a gap symbol (usually
“-”, but any other symbol may be defined in the descriptor) is that a
gap symbol represents an unspecified number of unknown symbols but “N”
stands for exatly one nucleic acid.
Amino acid symbols
|
|
|
| Name |
1-letter sybmol |
3-letter sybmol |
| Alanine |
A |
Ala |
| Arginine |
R |
Arg |
| Asparagine |
N |
Asn |
| Aspartic acid |
S |
Asp |
| Cysteine |
C |
Cys |
| Glutamic acid |
E |
Glu |
| Glutamine |
Q |
Gln |
| Glycine |
G |
Gly |
| Histidine |
H |
His |
| Isoleucine |
I |
Ile |
| Leucine |
L |
Leu |
| Lysine |
K |
Lys |
| Methionine |
M |
Met |
| Phenylalanine |
F |
Phe |
| Proline |
P |
Pro |
| Serine |
S |
Ser |
| Threonine |
T |
Thr |
| Tryptophan |
W |
Trp |
| Tyrosine |
Y |
Tyr |
| Valine |
V |
Val |
| Selenocysteine |
U |
Sec |
| Pyrrolysine |
O |
Pyl |
| Asparagine or aspartic acid |
B |
Asx |
| Glutamine or glutamic acid |
Z |
Glx |
| Leucine or Isoleucine |
J |
Xle |
| Unspecified or unknown amino acid |
X |
Xaa |
The symbols with grey background are
ambiguity symbols. The difference between “X” rsp. “Xaa” and a gap
symbol (e.g. “—”, but any other symbol may be defined in the
descriptor) is that a gap symbol represents an unspecified number of
unknown symbols but “X” rsp. “Xaa” stand for exatly one amino acid. The
amino acids “Selenocysteine” and Pyrrolysine" are non-standard amino
acids that only occur in certain species.
Continue with:
Edit Description
Resource links
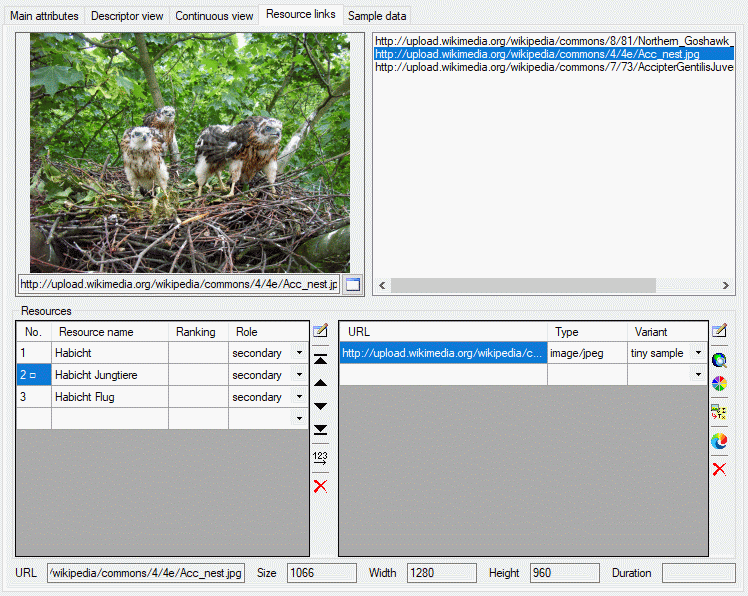
The resouce links tab allows assigment and inspection of resource links
for the description. In the lower Resources part of the tab there
are two tables. Ar the left the “Resource” table specifies the single
resources, at the right the “Resource variant” table contains the
associated URLs to images, video or sound resources.
To enter a new resource select the empty line at the end of the
“Resource” table and click on the “Resource name” field. After entering
the “Resource name” (leave the cell by pressing the TAB key)
automatically a new value for the sequence number (“No.”) is
initialized. The sequence number determines the display order in tables
and may be changed manually. You may order the resource entries by
clicking on the column header or by using the arrow buttons in the left
tool strip ( and
and  for up and down,
for up and down,
 and
and  to shift the
selected entry to top or bottom). After ordering the resources click
button
to shift the
selected entry to top or bottom). After ordering the resources click
button  to renumber the entries starting with “1”
and make the changes effective. In field “Ranking” you may enter a
numeric value in range 0 … 10 expressing the suitability of the
resource. In field “Role” you may select one of the offered values.
to renumber the entries starting with “1”
and make the changes effective. In field “Ranking” you may enter a
numeric value in range 0 … 10 expressing the suitability of the
resource. In field “Role” you may select one of the offered values.
If you want to delete a resource, you have three choices:
- Select the resource and press the
 delete button in
the left tool strip.
delete button in
the left tool strip.
- Select the resource and press the Delete key on the keyboard.
- Enter the sequence number column (“No.”) and remove the value. After
leaving the cell the entry will be deleted.
In any case you will be asked if you want to delete the data row, if
resource links are present.
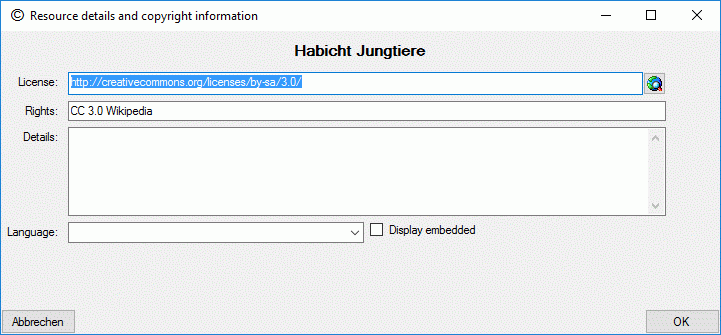
To view or edit the additional resource data and copyright information
press on the button  besides the resource
table. In field License: and a link to the license text in the web
may be entered, in field Rights: you may enter a copyright text. By
pressing the
besides the resource
table. In field License: and a link to the license text in the web
may be entered, in field Rights: you may enter a copyright text. By
pressing the  button a browser window can be
opened to navigate to the license page in the web. Furthermore you may
enter a detailled text (field Details:), the resource language
(field Language:) and set the Display embedded flag (see picture
below). If any additional resource data have been entered, this will be
indicated by a square (□) behind the resource name’s sequence number
(see picture above).
button a browser window can be
opened to navigate to the license page in the web. Furthermore you may
enter a detailled text (field Details:), the resource language
(field Language:) and set the Display embedded flag (see picture
below). If any additional resource data have been entered, this will be
indicated by a square (□) behind the resource name’s sequence number
(see picture above).
To enter a resource link, first select the entry in the “Resource”
table, then select the empty line at the end of the “Resource variant
table”. By clicking on the “URL” field a web address may be entered. By
double-clicking the “URL” field a browser window opens to navigate to
the resource. For each resource several URLs to resource variants with
different quality levels, e.g. different resolution, may be inserted.
For each entry in the URL table a different value of “Variant” must be
selected.
As an alternative to a resource URL a colour may be spcified in the
format “color://#rrggbb” where “rrggbb” specifies a hexadecimal colour
code. The colour can simply be seleced by clicking the button
 besides the resource variant table.
besides the resource variant table.
To remove a resource variant, select the entry and press the
 button in the right tool strip or the Delete key on the keyboard. Alternatively you
may delete the “URL” entry in the resource variant table.
button in the right tool strip or the Delete key on the keyboard. Alternatively you
may delete the “URL” entry in the resource variant table.
The button  besides the resource variant table
allows you to view a resource URL in the system browser.
besides the resource variant table
allows you to view a resource URL in the system browser.
The upper part of the resource links tab consists of a preview window on
the left side and the complete list of resource links. By selecting one
of the links the preview is shown on the preview window. If you press
button  in the preview window, the resource is
opened in a separate viewer window.
in the preview window, the resource is
opened in a separate viewer window.
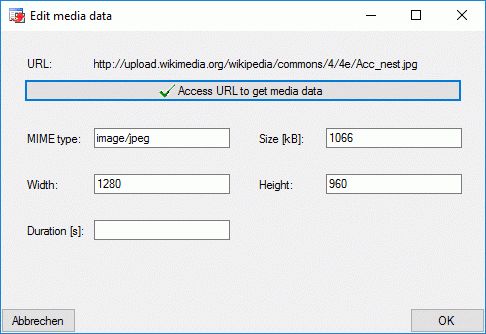
At the bottom of the resource links tab the media data of a selected URL
are displayed. These data can be edited by double-clicking the “Type”
entry in the resource variant table, by clicking at one of the media
data value fields or the  button. The edit
dialog provides the possibility to access the URL to get available data.
Additionally the values may be edited manually.
button. The edit
dialog provides the possibility to access the URL to get available data.
Additionally the values may be edited manually.
Copy description resources to a descriptor
If you have write access to descriptors of the currently selected
description’s project, you may copy a selected resource URL to a
descriptor. After clicking the  button a window
will open to select one of the accessible descriptors (see image
below). Remark: To see the copied resources in the Descriptorview or the Continuous
view, you will have to reconnect to
the database.
button a window
will open to select one of the accessible descriptors (see image
below). Remark: To see the copied resources in the Descriptorview or the Continuous
view, you will have to reconnect to
the database.
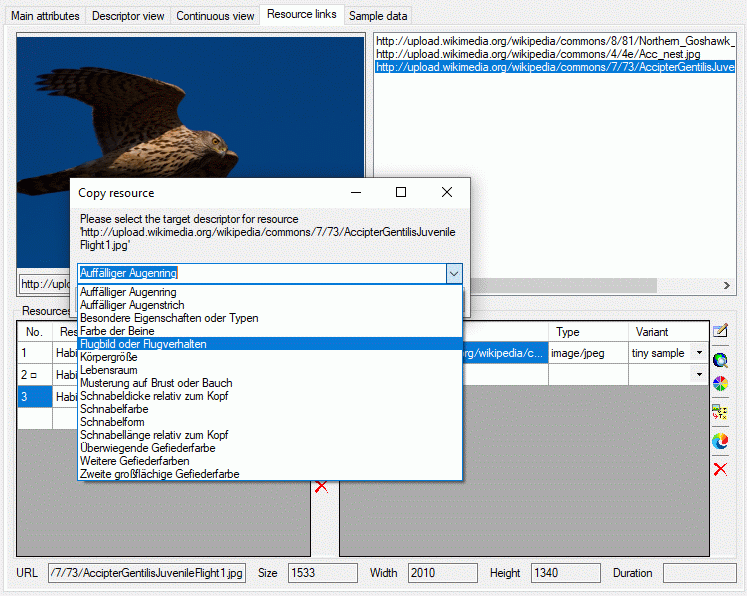
Continue with::
Edit Description
Sample data
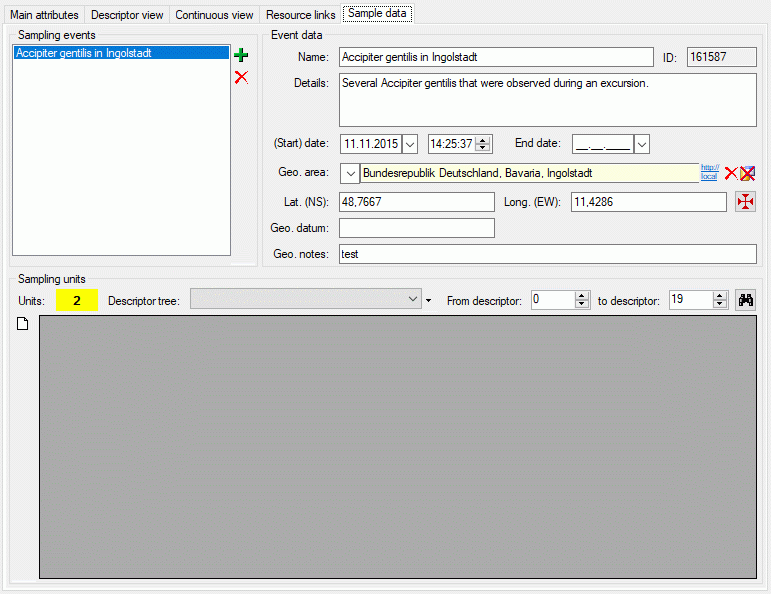
The sample data tab allows assigment and inspection of sampling events
and associated sampling units for the description. In the upper left
Sampling events part of the tab there is a list of the sampling
events stored for the description. By selecting a sampling event entry
the Event data are shown in the upper right part of the tab. Here
you may edit the event name, a detailled description, the date and time
rsp. a date and time span and geographic data. To insert a new sampling
event click on the  add button in the Sampling events
section, to delete the selected sampling event and all stored sampling
unit data press the
add button in the Sampling events
section, to delete the selected sampling event and all stored sampling
unit data press the  button (see image above).
button (see image above).
The geographic area may be linked to an entry of a DiversityGazetteers
database (see section Module related entry).
If latitiude and longituede have not yet been entered, the coordinates
of the selected DiversityGazetteers entry will be inserted. By pressing
the  button a window to select the
coordinates by Google maps will be opened. Field Geo. datum allows
entering short text concerning the geodetic datum. If coordinates are
entered using Google maps a remark that WGS84 coordinates are used will
be inserted.
button a window to select the
coordinates by Google maps will be opened. Field Geo. datum allows
entering short text concerning the geodetic datum. If coordinates are
entered using Google maps a remark that WGS84 coordinates are used will
be inserted.
Editing sampling unit data
For each sampling events one or several Sampling units may be
entered. A sampling unit represents a tuple of values that belong
together, e.g. because they represent a single specimen. For each
descriptor, which is represented by the table column, a value may be
entered in a sampling unit, which builds a single table line. The
background colour of each table column indicates the type of sample
data: categorical,
quantitative, text or sequence . If the number of sampling units is
displayed in field Units of the Sampling units section in the
lower part of the tab. If sampling units are present, the number is
displayed with yellow background (see
image below).
To view the unit data, select a Descriptor tree and choose the
descriptor sequence range (From descriptor … to descriptor) to
restrict the number of columns displayed in the unit table. Finally
press button  and the sampling untis will be
displayed (see image below). If you prefer to display the present
sampling units automatically whenever you select a sampling event, click
on the number behind Units. The background changes from yellow to red to indicate this mode. When you
select a cell and resources are available for the selected descriptor
column, the button
and the sampling untis will be
displayed (see image below). If you prefer to display the present
sampling units automatically whenever you select a sampling event, click
on the number behind Units. The background changes from yellow to red to indicate this mode. When you
select a cell and resources are available for the selected descriptor
column, the button  will be shown. You may click it
to view the descriptor resources.
will be shown. You may click it
to view the descriptor resources.
To insert a new sampling unit press button  . By
clicking on the free space at the beginning of a table line you mark the
whole sampling unit (see picture below). Now you have the additional
options to delete the unit by pressing button
. By
clicking on the free space at the beginning of a table line you mark the
whole sampling unit (see picture below). Now you have the additional
options to delete the unit by pressing button  or
to copy the data to a new sampling unit by pressing the
or
to copy the data to a new sampling unit by pressing the
 button. Keep in mind that all unit data will be
copied, even if you do not see all descriptor columns due to the display
restrictions.
button. Keep in mind that all unit data will be
copied, even if you do not see all descriptor columns due to the display
restrictions.
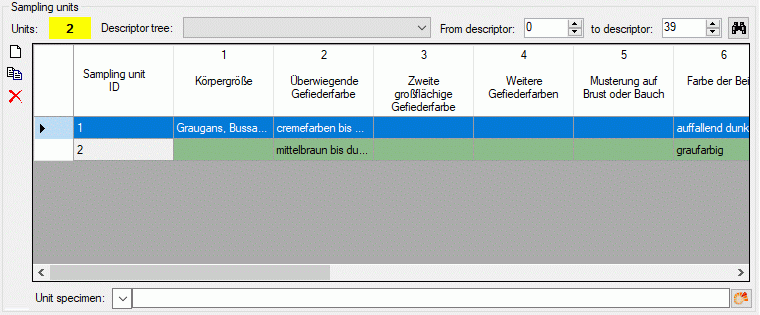
When you select the Sampling unit ID cell you may enter a collection
specimen in the Unit specimen control below the units table. By
clicking the  DWB button (see section
Module related entry) you can link it to an
entry in a DiversityCollection database. If the sampling unit is linked
to a database entry in DiversityCollection, the text of the Unit
specimen: control is shown with light
yellow background. You may view the linked data by clicking the
DWB button (see section
Module related entry) you can link it to an
entry in a DiversityCollection database. If the sampling unit is linked
to a database entry in DiversityCollection, the text of the Unit
specimen: control is shown with light
yellow background. You may view the linked data by clicking the
 button or double clicking the
Sampling unit ID cell.
button or double clicking the
Sampling unit ID cell.
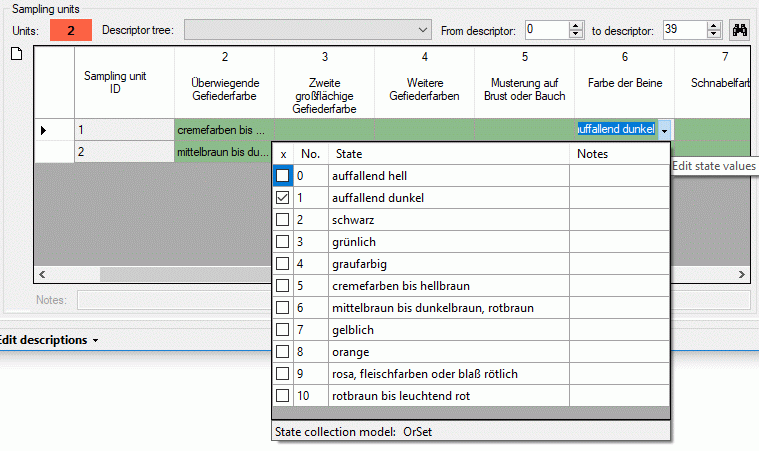
To modify categorical sampling data enter the table cell and press the
 button within the cell. A control will be
opened where you can select the categorical states and enter notes or
modifier values (if defined) for each single state (see image below).
button within the cell. A control will be
opened where you can select the categorical states and enter notes or
modifier values (if defined) for each single state (see image below).
For all other sampling data the value can be entered directly in the
table cell. Notes for the selected table cell are entered in the
Notes: text box directly below the units table. For categorical,
text and molecular sequence sample data a separate edit window can be
opened by double-clicking the table cell. For molecular sequence data
the tool strip buttons  (import) and
(import) and
 (export) allow import and export from rsp. to
dedicated file formats as described in sections Import sequencedata and Export sequence
data.
(export) allow import and export from rsp. to
dedicated file formats as described in sections Import sequencedata and Export sequence
data.
Continue with:






 to view the database link and
to view the database link and








 or view the data in a separate window by clicking
or view the data in a separate window by clicking ![]() . When you have collected all query results click the OK button and the scopes
are inserted for the description (see below).
. When you have collected all query results click the OK button and the scopes
are inserted for the description (see below).





 in the Project section (see pictures
above). A “read only” project cannot be selected for the actual
description. If the selected description has already been assigned to a
“read only” project by another user with appropriate rights, this is
indicated at the right bottom corner of the application window (see
above below right).
in the Project section (see pictures
above). A “read only” project cannot be selected for the actual
description. If the selected description has already been assigned to a
“read only” project by another user with appropriate rights, this is
indicated at the right bottom corner of the application window (see
above below right).





 button on the tool strip this example may be
hidden. Descriptors that are marked as mandatory are displayed with
button on the tool strip this example may be
hidden. Descriptors that are marked as mandatory are displayed with
 button the tree view may be
expanded to display the contained categorical states, quantitative
measures or texts. By pressing the
button the tree view may be
expanded to display the contained categorical states, quantitative
measures or texts. By pressing the  button the
tree will be collapsed to descriptor level.
button the
tree will be collapsed to descriptor level. , to delete a
descriptor select the entry and press
, to delete a
descriptor select the entry and press 
 (see
first entries in picture above). The resources may be accessed by a
right-click on the tree node and selecting context menu item
(see
first entries in picture above). The resources may be accessed by a
right-click on the tree node and selecting context menu item
 View resources (see picture above).
View resources (see picture above).

 opens an edit window at the right part
of the tab. This window provides the same functions as available in the
descriptor view tab. The picture below shows the resource linked to a
categorical state that can be opened by doule-clicking the state ID in
the Categorical states section.
opens an edit window at the right part
of the tab. This window provides the same functions as available in the
descriptor view tab. The picture below shows the resource linked to a
categorical state that can be opened by doule-clicking the state ID in
the Categorical states section. 










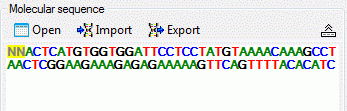

 Open →
Open → 









 button to
select the file where the data shall be exported. By default the data
are appended at the end of the selected file, check the Overwrite
existing file option to overwrite it. Finally click the
button to
select the file where the data shall be exported. By default the data
are appended at the end of the selected file, check the Overwrite
existing file option to overwrite it. Finally click the
 Save file button to write the data.
Save file button to write the data.
 Home to return to the HTML form.
Home to return to the HTML form. 
 Options you may
insert some additional input fields in the form, e.g. item details or
notes. With drop down button
Options you may
insert some additional input fields in the form, e.g. item details or
notes. With drop down button  Scope you may
insert input fields for the vasiour scope types and drop down button
Scope you may
insert input fields for the vasiour scope types and drop down button
 Color lets you select the background color.
All that changes come into affect, when you reopen the form or redraw
the HTML form using the
Color lets you select the background color.
All that changes come into affect, when you reopen the form or redraw
the HTML form using the  Reload button.
Reload button.

 for up and down,
for up and down,
 to renumber the entries starting with “1”
and make the changes effective. In field “Ranking” you may enter a
numeric value in range 0 … 10 expressing the suitability of the
resource. In field “Role” you may select one of the offered values.
to renumber the entries starting with “1”
and make the changes effective. In field “Ranking” you may enter a
numeric value in range 0 … 10 expressing the suitability of the
resource. In field “Role” you may select one of the offered values. besides the resource
table. In field License: and a link to the license text in the web
may be entered, in field Rights: you may enter a copyright text. By
pressing the
besides the resource
table. In field License: and a link to the license text in the web
may be entered, in field Rights: you may enter a copyright text. By
pressing the  button a browser window can be
opened to navigate to the license page in the web. Furthermore you may
enter a detailled text (field Details:), the resource language
(field Language:) and set the Display embedded flag (see picture
below). If any additional resource data have been entered, this will be
indicated by a square (□) behind the resource name’s sequence number
(see picture above).
button a browser window can be
opened to navigate to the license page in the web. Furthermore you may
enter a detailled text (field Details:), the resource language
(field Language:) and set the Display embedded flag (see picture
below). If any additional resource data have been entered, this will be
indicated by a square (□) behind the resource name’s sequence number
(see picture above).
 besides the resource variant table
allows you to view a resource URL in the system browser.
besides the resource variant table
allows you to view a resource URL in the system browser. in the preview window, the resource is
opened in a separate viewer window.
in the preview window, the resource is
opened in a separate viewer window.
 button a window
will open to select one of the accessible descriptors (see image
below). Remark: To see the copied resources in the
button a window
will open to select one of the accessible descriptors (see image
below). Remark: To see the copied resources in the 


 button a window to select the
coordinates by Google maps will be opened. Field Geo. datum allows
entering short text concerning the geodetic datum. If coordinates are
entered using Google maps a remark that WGS84 coordinates are used will
be inserted.
button a window to select the
coordinates by Google maps will be opened. Field Geo. datum allows
entering short text concerning the geodetic datum. If coordinates are
entered using Google maps a remark that WGS84 coordinates are used will
be inserted. and the sampling untis will be
displayed (see image below). If you prefer to display the present
sampling units automatically whenever you select a sampling event, click
on the number behind Units. The background changes from
and the sampling untis will be
displayed (see image below). If you prefer to display the present
sampling units automatically whenever you select a sampling event, click
on the number behind Units. The background changes from  button. Keep in mind that all unit data will be
copied, even if you do not see all descriptor columns due to the display
restrictions.
button. Keep in mind that all unit data will be
copied, even if you do not see all descriptor columns due to the display
restrictions.
 button within the cell. A control will be
opened where you can select the categorical states and enter notes or
modifier values (if defined) for each single state (see image below).
button within the cell. A control will be
opened where you can select the categorical states and enter notes or
modifier values (if defined) for each single state (see image below).