Diversity TaxonNames
TABLES, VIEWS, FUNCTIONS, PROCEDURES, ROLES
The following objects are not included:
- Logging tables
- Enumeration tables
- System objects
- Objects marked as obsolete
- Previous versions of objects
TABLES
Table ProjectUser
User with access to a project
| Column |
Data type |
Description |
Nullable |
Relation |
| LoginName |
nvarchar (50) |
ID of the User as stored in table UserProxy |
NO |
- |
| ProjectID |
int |
ID of the project. Foreign key, relates to PK of table ProjectProxy |
NO |
Refers to table ProjectProxy |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
Table ReplicationPublisher
Source providing data for replication
| Column |
Data type |
Description |
Nullable |
Relation |
| DatabaseName |
varchar (255) |
The name of the publishing database |
NO |
- |
| Server |
varchar (255) |
The name or address of the server where the publishing database is located |
NO |
- |
| Port |
smallint |
The port used by the server |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: user_name() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: user_name() |
YES |
- |
Table SynonymyListCache
Storing results of function SynonymyList
| Column |
Data type |
Description |
Nullable |
Relation |
| ID |
int |
NameID of the selected taxon |
NO |
- |
| ProjectID |
int |
ProjectID of the synonymy |
NO |
- |
| NameID |
int |
ID of the synonymized name. Refers to the NameID of TaxonName (= foreign key). |
NO |
- |
| DisplayText |
nvarchar (900) |
A preformatet textstring for display in a synoymlist |
YES |
- |
| TaxonName |
nvarchar (500) |
The name of the taxon including the genus and the species epithet |
YES |
- |
| TaxonomicRank |
nvarchar (50) |
Taxonomic rank of the taxon (var., subsp., species, genus, family, order, etc.). The rank must be selected from the associated list of ranks (= TaxonomyRank) |
YES |
- |
| InfraspecificEpithet |
nvarchar (200) |
The epithet of the infraspecific entity |
YES |
- |
| Authors |
nvarchar (500) |
The combined authors of this name |
YES |
- |
| SynonymText |
nvarchar (500) |
The text string containing the citation of the synonymisation |
YES |
- |
| Prefix |
nvarchar (5) |
The prefix as used in synonym lists |
YES |
- |
| Ord |
int |
The preferred sequence of the synonyms. The numbers in Ord correspond to the different kinds of the synonyms: |
|
|
| -20 = Accepted name |
|
|
|
|
| -10 = Basionym |
|
|
|
|
| 0 = Homotypic Synonym |
|
|
|
|
0 = Heterotypic Synoyms
x + 1 = Isonym resp. Assigned to like ‘duplicate’, ‘isonym’, ‘orthographic variant’ (will be display after the name it is a variant of)| YES| -|
| YearOfPubl| int| The year when the name was published| YES| -|
| TaxonNameCache| nvarchar (255)| A full unique version of the name. Generated by the database, not entered by the user (candidate key)| YES| -|
| ConceptSuffix| nvarchar (200)| Empty for default and primary concept; else ’s. lat.’, ’s. str.’, ‘sec.’, ‘sensu Muell.’, ’emend.’ , ‘(Auct.)’ etc.| YES| -|
| ConceptNotes| nvarchar (500)| Notes on the concept of the name, e.g. incl. forms with black apothecia| YES| -|
| BasedOnNameID| int| The Basionym of this name, resp. the NameID of the Basionym. A name is a Basionym, if NameID = BasionymID.| YES| -|
| SynType| nvarchar (50)| Type of the synonymization, e.g. ‘heterotypic’, ‘homotypic’| YES| -|
| SynNameID| int| The ID of the synonym to which this name has been assigned to. Refers to the NameID of TaxonName (= foreign key).| NO| -|
| NameParentID| int| The NameID of the parent within the synonymy| YES| -|
Table TaxonAcceptedName
TaxonAcceptedName contains the accepted names (“valid or preferred taxon names”). For each project, a name in “TaxonName” may either be accepted (a corresponding record in “TaxonAcceptedName” exists) or not. Examples for not accepted names are synonyms of accepted names and nomenclaturally rejected names.
| Column |
Data type |
Description |
Nullable |
Relation |
| ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module.Default value: 0 |
NO |
Refers to table ProjectProxy and table TaxonNameProject |
| NameID |
int |
ID of the accepted name. Refers to the NameID of TaxonName (= foreign key). |
NO |
Refers to table TaxonNameProject |
| IgnoreButKeepForReference |
tinyint |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead.Default value: 0 |
NO |
- |
| ConceptSuffix |
nvarchar (200) |
Empty for default and primary concept; else ’s. lat.’, ’s. str.’, ‘sec.’, ‘sensu Muell.’, ’emend.’ , ‘(Auct.)’ etc.Default value: '’ |
YES |
- |
| ConceptNotes |
nvarchar (500) |
Notes on the concept of the name, e.g. incl. forms with black apothecia |
YES |
- |
| RefURI |
varchar (255) |
ReferenceURI: Source publication where synonymization is published (not publication of name!) |
YES |
- |
| RefText |
nvarchar (255) |
Free text, esp. where a RefURI is missing. Source publication where synonymization is published (not publication of name!) |
YES |
- |
| RefDetail |
nvarchar (255) |
Esp. page number on which the synonymization is published |
YES |
- |
| TypistsNotes |
nvarchar (255) |
An internal note of the responsible person concerning this synonymization. This information is NOT included in any report. |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data. This is the operator (or typist) name, which may be different from the person responsible.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when record was first entered (typed or imported) into this system.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data. This is the operator (or typist) name, which may be different from the person responsible.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when record was last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
- ProjectProxy
- TaxonNameProject
trgInsTaxonAcceptedName
Setting version of taxon
Table TaxonCommonName
TaxonCommonName contains the common names that are assigned to a scientific name.
| Column |
Data type |
Description |
Nullable |
Relation |
| NameID |
int |
ID taxonomic name. Refers to the column NameID of the table TaxonName (= foreign key). |
NO |
Refers to table TaxonName |
| CommonName |
nvarchar (220) |
A common name of the taxonomic name |
NO |
- |
| LanguageCode |
varchar (2) |
The 2-letter code of the language of the common name according to ISO |
NO |
Refers to table LanguageCode_Enum |
| CountryCode |
varchar (2) |
The 2-letter code of the country where the common name is used according to ISO ISO 3166-1 |
NO |
Refers to table CountryCode_Enum |
| ReferenceTitle |
nvarchar (220) |
The title of the reference where the common name was published |
NO |
- |
| ReferenceURI |
varchar (255) |
The URI of the reference e.g. as provided by the module DiversityReferences |
YES |
- |
| ReferenceDetails |
nvarchar (500) |
Esp. page number on which the common name is published |
YES |
- |
| SubjectContext |
nvarchar (500) |
The context in which the common name is used, e.g. pharmacy, food |
YES |
- |
| Transliteration |
nvarchar (255) |
Transliteration of the original name |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes about the common name |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Who inserted this datasetDefault value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
The time when this dataset was insertedDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Who was the last to update this datasetDefault value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
The last time when this dataset was updatedDefault value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
- CountryCode_Enum
- LanguageCode_Enum
- TaxonName
trgInsTaxonCommonName
Setting version of taxon
Table TaxonGeography
TaxonGeography is containing the information about the geographic distribution according to the protologue.
| Column |
Data type |
Description |
Nullable |
Relation |
| NameID |
int |
ID of the name. Refers to the NameID of TaxonName (= foreign key). |
NO |
Refers to table TaxonName |
| PlaceURI |
varchar (255) |
URI (e.g. LSID) within e.g. the DiversityGazetteer for a place (which may have several names) as found in the protologue |
NO |
- |
| PlaceNameCache |
nvarchar (100) |
The name of the place |
NO |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when the data were first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when the data were last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
trgInsTaxonGeography
Setting version of taxon
Table TaxonHierarchy
TaxonHierarchy contains information about the hierarchical position of the taxa. For each project, a name may be referred to a single parent taxon. The attribute “NameParentID” may, however, be Null, in which case “HierarchyPositionIsUncertain” should be true.
| Column |
Data type |
Description |
Nullable |
Relation |
| ProjectID |
int |
Each project can implement a different taxonomic hierarchy. Refers to the common project definition in the DiversityProjects module.Default value: 0 |
NO |
Refers to table ProjectProxy and table TaxonNameProject |
| NameID |
int |
Unique NameID code of the higher taxon. Refers to the NameID code of LichenName (= foreign key). |
NO |
Refers to table TaxonNameProject |
| IgnoreButKeepForReference |
tinyint |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead.Default value: 0 |
NO |
- |
| NameParentID |
int |
Next higher taxon (e.g. the family or subfamily if this taxon is a genus) |
YES |
- |
| HierarchyRefURI |
varchar (255) |
Reference URI: Source publication where the hierarchy is published (not publication of name!) as stored e.g. in DiversityReferences |
YES |
- |
| HierarchyRefText |
nvarchar (255) |
Free citation, esp. where a HierarchyRefURI is missing. Source publication where the hierarchy is published (not publication of name!) |
YES |
- |
| HierarchyRefDetail |
nvarchar (255) |
Esp. page number on which the hierarchy is published |
YES |
- |
| HierarchyPositionIsUncertain |
bit |
If the position of this taxon within this taxonomic group is uncertain.Default value: 0 |
YES |
- |
| HierarchyTypistsNotes |
nvarchar (255) |
An internal note of the responsible person concerning this hierarchy. This information is NOT included in any report. |
YES |
- |
| HierarchyListCache |
nvarchar (1000) |
CALCULATED FIELD: List of higher taxa for faster access: “div.; class; ord.; fam.” |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data. This is the operator (or typist) name, which may be different from the person responsible.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when record was first entered (typed or imported) into this system.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data. This is the operator (or typist) name, which may be different from the person responsible.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when record was last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
- ProjectProxy
- TaxonNameProject
trgInsTaxonHierarchy
Setting version of taxon and HierarchyListCache
Table TaxonName
TaxonName is the nomenclatural information. Most items are expected to be undisputed nomenclatural facts. Some data items include editorial opinion (nomenclatural status, but also the accepted spelling which may involve orthographical or grammatical corrections as required by ICBN). However, it is expected that within a project agreement can be achieved on the information contained in TaxonName, i.e. collaborators can “correct” information without consultation.
| Column |
Data type |
Description |
Nullable |
Relation |
| NameID |
int |
The ID of a name in DiversityTaxonNames_Fungi (primary key) |
NO |
- |
| TaxonNameCache |
nvarchar (255) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
YES |
- |
| Version |
smallint |
The version of a name record (revision number, internally filled by system)Default value: (1) |
YES |
- |
| BasedOnNameID |
int |
The Basionym of this name, resp. the NameID of the Basionym. A name is a Basionym, if NameID = BasionymID. |
YES |
Refers to table TaxonName |
| CreationType |
nvarchar (50) |
E.g.: taxon based on new type, combination based on a previously publ. name (‘comb. nov.’), new name (’nom.nov.’) introduced to replace a homonym (may occur for genera!), validation of previously invalidly publ. taxon name (’ex’), or unknown. |
YES |
Refers to table TaxonNameCreationType_Enum |
| TaxonomicRank |
nvarchar (50) |
Taxonomic rank of the taxon (var., subsp., species, genus, family, order, etc.). The rank must be selected from the associated list of ranks (= TaxonomyRank)Default value: N’sp.’ |
NO |
Refers to table TaxonNameTaxonomicRank_Enum |
| GenusOrSupragenericName |
nvarchar (200) |
If rank is above species: Name of taxon above species level (currently accepted spelling). Includes infrageneric taxon names, genera, families, etc. |
NO |
- |
| SpeciesGenusNameID |
int |
If rank is species or below: NameID of the Genus name. Refers to same table with TaxonomicRank = genus. |
YES |
Refers to table TaxonName |
| InfragenericEpithet |
nvarchar (200) |
If rank below genus and above species: Name of infrageneric taxon above species level (currently accepted spelling). Includes subgenus, series, etc. |
YES |
- |
| SpeciesEpithet |
nvarchar (100) |
The species name part of the species name, for example ‘alba’ in ‘Abies alba’. |
YES |
- |
| InfraspecificEpithet |
nvarchar (100) |
The epithet of the infraspecific entity |
YES |
- |
| BasionymAuthors |
nvarchar (100) |
The authors of a newly created name. For all taxonomic ranks, only for ‘comb. nov.’ or ’nom. nov.’: Author(s) of the basionym (will be displayed in ‘()’, do not enter the parentheses), abbreviated according to authors standard |
YES |
- |
| CombiningAuthors |
nvarchar (255) |
The names of the combining authors if the name is base on another older name (e.g. combined into a different genus) |
YES |
- |
| PublishingAuthors |
nvarchar (255) |
If the authors of the taxon differ from the authors of the publication: the latter (…‘in ’ Publishing authors … ) |
YES |
- |
| SanctioningAuthor |
nvarchar (100) |
Sanctioning is a special instrument available for fungi to allow the starting date of taxonomy to be identical with higher plants, but preserve names used by Fries (Fr.) and Person (Pers.) |
YES |
- |
| NonNomenclaturalNameSuffix |
nvarchar (200) |
A suffix for concept names not included in any nomenclatural code. If present fields for authors and typification must be empty and NomenclaturalCode set to ’not under code’. |
YES |
- |
| IsRecombination |
bit |
True if the name is a recombinationDefault value: (0) |
NO |
- |
| IsHybrid |
bit |
If checked (or ‘True’), the new taxon is a hybrid with or without a hybrid epithet.Default value: (0) |
YES |
- |
| HybridNameID1 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 1. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
Refers to table TaxonName |
| HybridNameID2 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 2. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
Refers to table TaxonName |
| HybridNameID3 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 3. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
Refers to table TaxonName |
| HybridNameID4 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 4. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
Refers to table TaxonName |
| ReferenceTitle |
nvarchar (600) |
The title of the publication where the name was published. Note this is only a cached value where ReferenceURI is present |
YES |
- |
| ReferenceURI |
varchar (255) |
URI (e.g. LSID) of Reference, referes to table ReferenceTitle in Database DiversityReferences: Source publication where name is published |
YES |
- |
| Volume |
nvarchar (20) |
The volume of the journal |
YES |
- |
| Issue |
nvarchar (255) |
The issue of the literature |
YES |
- |
| Pages |
nvarchar (50) |
The pages within the literature |
YES |
- |
| DetailLocation |
nvarchar (200) |
Additional information like plates etc. |
YES |
- |
| DayOfPubl |
tinyint |
The day when the name was published |
YES |
- |
| MonthOfPubl |
tinyint |
The month when the name was published |
YES |
- |
| YearOfPubl |
smallint |
The year when the name was published |
YES |
- |
| DateOfPublSupplement |
nvarchar (200) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
YES |
- |
| YearOnPubl |
smallint |
The year cited on the original paper as year of publication |
YES |
- |
| DateOnPublSupplement |
nvarchar (200) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
YES |
- |
| Protologue |
nvarchar (MAX) |
Full text of the protologue. If protologue is provided in Latin and in another language |
|
|
| languages, both only the Latin or several languages may be stored together. |
YES |
- |
|
|
| ProtologueURI |
varchar (255) |
URI of externally available Protologue information (scanned image, full text from external provider, etc.) |
YES |
- |
| ProtologueResourceURI |
varchar (255) |
The ResourceID of an image of the protologue as stored in the module DiversityResources. Primary key of table Resource in the database DiversityResources.(= Foreign key) |
YES |
- |
| NameUsageReferences |
nvarchar (255) |
Indexing volumes like Index of Fungi or any Name usage that caused this name to be added to the database |
|
|
| YES |
- |
|
|
|
| OriginalOrthography |
nvarchar (255) |
The original spelling of the name |
YES |
- |
| NomenclaturalCode |
nvarchar (50) |
Code of Nomenclature under which this taxon was created: ‘Bacteriology’, ‘Botany’ (incl. Mycology), ‘Zoology’, ‘Biocode’ (for future use), ‘Non nomenclatural name’. Default value: ‘Botany’ |
YES |
Refers to table TaxonNameNomenclaturalCode_Enum |
| NomenclaturalStatus |
nvarchar (50) |
Categories for effective/valid/legitimate… esp. ’nom. illeg.’, ’nom. inval.’, ’nom. nudum’, etc. |
YES |
Refers to table TaxonNameNomenclaturalStatus_Enum |
| NomenclaturalComment |
nvarchar (MAX) |
Comments on the nomenclature. e.g. ‘according to ICBN Art. 39.1’ |
YES |
- |
| Typification |
nvarchar (50) |
The status of the type specimen(s) as written in the protologue, e.g. holotype |
YES |
- |
| TypificationDetails |
nvarchar (255) |
Details concerning the typification as written in the protologue, e.g. if just parts of a specimen were accepted as the type |
YES |
- |
| TypificationReferenceTitle |
nvarchar (255) |
The title of the publication where the typification was published. Note this is only a cached value where Typification-LiteratureLink is present |
YES |
- |
| TypificationReferenceURI |
varchar (255) |
URI (e.g. LSID) of Reference, refers to e.g DiversityReferences: Source publication where the typification is published |
YES |
- |
| TypificationNotes |
nvarchar (200) |
Notes concerning the typification |
YES |
- |
| TypeSubstrate |
nvarchar (255) |
The substrate the type was growing on as written in the protologue |
YES |
- |
| TypeLocality |
nvarchar (255) |
The locality where the type was found as written in the protologue |
YES |
- |
| TypeSpecimenNotes |
nvarchar (200) |
Notes about the type specimen, e.g. the herbarium where a type specimen is located, Collector, collection date/number, etc. |
YES |
- |
| AnamorphTeleomorph |
nvarchar (255) |
If the name is related to a anamorph or a teleomorph as written in the protologue |
YES |
- |
| TypistNotes |
nvarchar (200) |
Additional notes and problems |
YES |
- |
| RevisionLevel |
nvarchar (50) |
The level of the revision of the taxonomic name, e.g. ‘unchecked’, ‘fully checked’ |
YES |
Refers to table TaxonNameRevisionLevel_Enum |
| IgnoreButKeepForReference |
bit |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead.Default value: (0) |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the dataset is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data. This is the operator (or typist) name, which may be different from the person responsible.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when record was first entered (typed or imported) into this system.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data. This is the operator (or typist) name, which may be different from the person responsible.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when record was last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
| BasionymAuthorsYear |
int |
The year when the name was published by the authors of the Basionym |
YES |
- |
Depending on:
- TaxonNameCreationType_Enum
- TaxonNameNomenclaturalCode_Enum
- TaxonNameNomenclaturalStatus_Enum
- TaxonNameRevisionLevel_Enum
- TaxonNameTaxonomicRank_Enum
trgInsTaxonName
Setting TaxonNameCache
Table TaxonNameExternalDatabase
TaxonNameExternalDatabase document the sources of the names.
| Column |
Data type |
Description |
Nullable |
Relation |
| ExternalDatabaseID |
int |
An ID to identify an external data collection of plant names (primary key, the ID has no meaning outside of the DiversityWorkbench system) |
NO |
- |
| ExternalDatabaseName |
nvarchar (800) |
The name of the data collection that has been integrated or can be linked to for further analysis |
NO |
- |
| ExternalDatabaseVersion |
nvarchar (255) |
The version of this data collection (either official version number, or dates when the collection was integrated) |
YES |
- |
| Rights |
nvarchar (500) |
A description of copyright agreements or permission to use data from the external database |
YES |
- |
| ExternalDatabaseAuthors |
nvarchar (200) |
The persons or institutions responsible for the external database |
YES |
- |
| ExternalDatabaseURI |
nvarchar (300) |
The URI of the database provider resp. the external database |
YES |
- |
| ExternalDatabaseInstitution |
nvarchar (300) |
The institution responsible for the external database |
YES |
- |
| InternalNotes |
nvarchar (1500) |
Additional notes concerning this data collection |
YES |
- |
| ExternalAttribute_NameID |
nvarchar (255) |
The table and field name in the external data collection to which TaxonNameExternalID refers |
YES |
- |
| PreferredSequence |
tinyint |
For selection in e.g. picklists: of several equal names only the name from the source with the lowest preferred sequence will be provided. |
YES |
- |
| Disabled |
bit |
If this source should be disabled for selection of names e.g. in picklists |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data.Default value: user_name() |
YES |
- |
| LogCreatedWhen |
datetime |
Date and time when the data were first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
datetime |
Date and time when the data were last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Table TaxonNameExternalID
TaxonNameExternalID document the source of a name together with the ID of the source.
| Column |
Data type |
Description |
Nullable |
Relation |
| NameID |
int |
The ID of a name in DiversityTaxonNames (foreign key + part of primary key: the name string associated with NameID may occur in multiple external databases) |
NO |
Refers to table TaxonName |
| ExternalDatabaseID |
int |
The ID of an external taxonomic data collection as defined in TaxontNameExternalDatabase (foreign key + part of primary key) |
NO |
Refers to table TaxonNameExternalDatabase |
| ExternalNameURI |
varchar (255) |
The URI (e.g. a LSID) of the external name as defined in the external database |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data.Default value: user_name() |
YES |
- |
| LogCreatedWhen |
datetime |
Date and time when the data were first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
datetime |
Date and time when the data were last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
- TaxonName
- TaxonNameExternalDatabase
Table TaxonNameList
The table is containing the names listed in e.g. a checklist corresponding to the project.
| Column |
Data type |
Description |
Nullable |
Relation |
| NameID |
int |
ID of the name. Refers to the NameID of TaxonName (= foreign key). |
NO |
Refers to table TaxonName |
| ProjectID |
int |
Each project can may contain one taxon list. Refers to the common project definition in the DiversityProjects module. |
NO |
Refers to table TaxonNameListProjectProxy |
| Notes |
nvarchar (MAX) |
An note concerning this entry. |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when the data were first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when the data were last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
- TaxonName
- TaxonNameListProjectProxy
Table TaxonNameListAnalysis
Analysis values for list entries in the database, “Red list category: R”, “Time of observation: Sept. - Nov.”
| Column |
Data type |
Description |
Nullable |
Relation |
| NameID |
int |
ID of the name. Refers to the NameID of TaxonName (= foreign key). |
NO |
Refers to table TaxonNameList and table TaxonNameListReference |
| ProjectID |
int |
Each project can may contain one taxon list. Refers to the common project definition in the DiversityProjects module. |
NO |
Refers to table TaxonNameList and table TaxonNameListReference |
| TaxonNameListRefID |
int |
Unique ID of the reference within the list, refers to table TaxonNameListReference, part of primary keyDefault value: (-1) |
NO |
Refers to table TaxonNameListReference |
| AnalysisID |
int |
ID of the analysis (= foreign key) |
NO |
Refers to table TaxonNameListAnalysisCategory |
| AnalysisValue |
nvarchar (MAX) |
The result of the analysis |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes concerning this analysis |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when the data were first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when the data were last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
- TaxonNameList
- TaxonNameListAnalysisCategory
- TaxonNameListReference
Table TaxonNameListAnalysisCategory
Analysis types used within the database, e.g. “Red list category”, “Frequency”
| Column |
Data type |
Description |
Nullable |
Relation |
| AnalysisID |
int |
ID of the analysis (Primary key) |
NO |
- |
| AnalysisParentID |
int |
Analysis ID of the parent analysis if it belongs to a certain type documented in this table |
YES |
- |
| DisplayText |
nvarchar (50) |
Name of the analysis as e.g. shown in user interface |
YES |
- |
| Description |
nvarchar (MAX) |
Description of the analysis |
YES |
- |
| AnalysisURI |
varchar (255) |
URI referring to an external documentation of the analysis |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes concerning this analysis |
YES |
- |
| LogCreatedWhen |
datetime |
The time when this dataset was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Who created this datasetDefault value: user_name() |
YES |
- |
| LogUpdatedWhen |
datetime |
The last time when this dataset was updatedDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Who was the last to update this datasetDefault value: user_name() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
| ReferenceTitle |
nvarchar (800) |
The title of the publication where details about the analysis are published. Note this is only a cached value where ReferenceURI is present |
YES |
- |
| ReferenceURI |
varchar (400) |
URI (e.g. LSID) of Reference, referes to table ReferenceTitle in module DiversityReferences: Source publication where details about the analysis are published |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| SortingID |
int |
An ID for sorting the data |
YES |
- |
Table TaxonNameListAnalysisCategoryValue
Value lists for analysis types with predefined values, e.g. “0, 1, 2, 3, …” for Red list category. Includes description etc. for the values in the list.
| Column |
Data type |
Description |
Nullable |
Relation |
| AnalysisID |
int |
ID of the analysis (Primary key) |
NO |
Refers to table TaxonNameListAnalysisCategory |
| AnalysisValue |
nvarchar (255) |
The categorized value of the analysis |
NO |
- |
| Description |
nvarchar (500) |
Description of enumerated object, displayed in the user interface |
YES |
- |
| DisplayText |
nvarchar (50) |
Short abbreviated description of the object, displayed in the user interface |
YES |
- |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
YES |
- |
| Notes |
nvarchar (500) |
Internal development notes about usage, definition, etc. of an enumerated object |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when the data were first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when the data were last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
- TaxonNameListAnalysisCategory
Table TaxonNameListArea
The area defined for the whole taxon list
| Column |
Data type |
Description |
Nullable |
Relation |
| ProjectID |
int |
Each project can have a different opinion regarding the parameters defined for a list. Refers to the common project definition in the DiversityProjects module. |
NO |
Refers to table TaxonNameListProjectProxy |
| PlaceURI |
varchar (255) |
URI or identifier for a place (which may have several names) derived from e.g. TDWG |
NO |
- |
| PlaceNameCache |
nvarchar (255) |
The name of the place |
YES |
- |
| Notes |
nvarchar (MAX) |
An note concerning this entry. |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when the data were first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when the data were last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
- TaxonNameListProjectProxy
Table TaxonNameListCollectionSpecimen
The collection specimens on which the taxon list is based
| Column |
Data type |
Description |
Nullable |
Relation |
| NameID |
int |
ID of the name. Refers to the NameID of TaxonName (= foreign key). |
NO |
Refers to table TaxonNameList |
| ProjectID |
int |
ProjectID of the taxon list. Foreign key and part of primary key. |
NO |
Refers to table TaxonNameList |
| DisplayText |
varchar (255) |
The name of the collection specimen as shown e.g. in a user interface. Part of the primary key |
NO |
- |
| CollectionSpecimenURI |
varchar (255) |
URI of the collection specimen. May refer to the module CollectionSpecimen |
YES |
- |
| Role |
nvarchar (50) |
The role of the specimen in relation to the taxon list (= foreign key, see table TaxonNameListSpecimenRole_Enum) |
YES |
Refers to table TaxonNameListSpecimenRole_Enum |
| Description |
nvarchar (MAX) |
Description of the collection specimen |
YES |
- |
| PlaceURI |
varchar (255) |
URI or identifier for a place (which may have several names) derived from e.g. TDWG |
YES |
- |
| PlaceNameCache |
nvarchar (255) |
The name of the place |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes about the collection specimen |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when the data were first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when the data were last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
- TaxonNameList
- TaxonNameListSpecimenRole_Enum
Table TaxonNameListDistribution
The geographical distrbution for the organisms within the taxon list.
| Column |
Data type |
Description |
Nullable |
Relation |
| NameID |
int |
ID of the name. Refers to the NameID of TaxonName (= foreign key). |
NO |
Refers to table TaxonNameList and table TaxonNameListReference |
| ProjectID |
int |
Each project can may contain one taxon list. Refers to the common project definition in the DiversityProjects module. |
NO |
Refers to table TaxonNameList and table TaxonNameListReference |
| TaxonNameListRefID |
int |
Unique ID of the reference within the list, refers to table TaxonNameListReference, part of primary keyDefault value: (-1) |
NO |
Refers to table TaxonNameListReference |
| PlaceURI |
varchar (255) |
URI or identifier for a place (which may have several names) derived from e.g. TDWGDefault value: '’ |
NO |
- |
| PlaceNameCache |
nvarchar (255) |
The name of the placeDefault value: '’ |
NO |
- |
| Notes |
nvarchar (MAX) |
Notes concerning this analysis |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when the data were first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when the data were last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
- TaxonNameList
- TaxonNameListReference
Table TaxonNameListImage
Media, e.g. images of the organisms within the taxon list
| Column |
Data type |
Description |
Nullable |
Relation |
| NameID |
int |
ID of the accepted name. Refers to the NameID of TaxonName (= foreign key). |
NO |
Refers to table TaxonNameList |
| ProjectID |
int |
Each project can may contain one taxon list. Refers to the common project definition in the DiversityProjects module. |
NO |
Refers to table TaxonNameList |
| URI |
varchar (255) |
The complete URI address of the image. |
NO |
- |
| Description |
xml (MAX) |
Description of the image |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes concerning this analysis |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when the data were first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when the data were last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
Table TaxonNameListReference
The reference where the taxon list resp. the name is published
| Column |
Data type |
Description |
Nullable |
Relation |
| NameID |
int |
ID of the name. Refers to the NameID of TaxonName (= foreign key). |
NO |
Refers to table TaxonNameList |
| ProjectID |
int |
Each project can may contain one taxon list. Refers to the common project definition in the DiversityProjects module. |
NO |
Refers to table TaxonNameList |
| TaxonNameListRefID |
int |
Unique ID of the reference within the list, part of primary key |
NO |
- |
| TaxonNameListRefText |
nvarchar (800) |
Free text, esp. where a TaxonNameListRefURI is missing. Source publication where distribution is published (not publication of name!) |
NO |
- |
| TaxonNameListRefURI |
varchar (255) |
Reference URI: Source publication where the distribution is published (not publication of name!) as stored e.g. in DiversityReferences |
YES |
- |
| TaxonNameListRefDetail |
nvarchar (255) |
Esp. page number on which the distribution is published |
YES |
- |
| ReferenceType |
nvarchar (50) |
The type of the reference, e.g. a webpage |
YES |
Refers to table TaxonNameListReferenceType_Enum |
| TaxonUsageContext |
nvarchar (50) |
The context of the usage of the taxonomic name within the reference, e.g. ecology |
YES |
- |
| Notes |
nvarchar (MAX) |
An note concerning this entry. |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when the data were first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when the data were last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
- TaxonNameList
- TaxonNameListReferenceType_Enum
Table TaxonNameListUser
The table is containing the lists accessible for a user.
| Column |
Data type |
Description |
Nullable |
Relation |
| LoginName |
nvarchar (50) |
Login of a user. |
NO |
- |
| ProjectID |
int |
Project accessible for a user. Refers to ProjectID in table TaxonNameList (= foreign key). |
NO |
Refers to table TaxonNameListProjectProxy |
Depending on:
- TaxonNameListProjectProxy
Table TaxonNameProject
TaxonNameProject documents the projects of the names and provides access to the data.
| Column |
Data type |
Description |
Nullable |
Relation |
| NameID |
int |
The ID of a name in DiversityTaxonNames (foreign key + part of primary key: the name may occur in multiple projects) |
NO |
Refers to table TaxonName |
| ProjectID |
int |
ID of the project. Refers to the common project definition in the DiversityProjects module.Default value: 0 |
NO |
Refers to table ProjectProxy |
| LogInsertedWhen |
datetime |
The time when this dataset was insertedDefault value: getdate() |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Who inserted this datasetDefault value: user_name() |
YES |
- |
| LogUpdatedWhen |
datetime |
The last time when this dataset was updatedDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Who was the last to update this datasetDefault value: user_name() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
Table TaxonNameReference
References related to a taxonomic name
| Column |
Data type |
Description |
Nullable |
Relation |
| NameID |
int |
ID of the name. Refers to the NameID of TaxonName (= foreign key). |
NO |
Refers to table TaxonName |
| TaxonNameRefID |
int |
Unique ID of the reference, part of primary key |
NO |
- |
| TaxonNameRefText |
nvarchar (800) |
The citation text of the reference |
NO |
- |
| TaxonNameRefURI |
varchar (500) |
Reference URI: Link to the source where the publication as stored e.g. DiversityReferences |
YES |
- |
| TaxonNameRefDetail |
nvarchar (255) |
Esp. page number on which the distribution is published |
YES |
- |
| ReferenceType |
nvarchar (50) |
The type of the reference, e.g. a webpage |
YES |
Refers to table TaxonNameListReferenceType_Enum |
| Notes |
nvarchar (MAX) |
An note concerning this entry. |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when the data were first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when the data were last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
- TaxonName
- TaxonNameListReferenceType_Enum
Table TaxonNameResource
Resouces related to the taxonomic name
| Column |
Data type |
Description |
Nullable |
Relation |
| NameID |
int |
ID of the name. Refers to the NameID of TaxonName (= foreign key). |
NO |
Refers to table TaxonNameProject |
| ProjectID |
int |
ID of the project. Refers to the common project definition in the DiversityProjects module. |
NO |
Refers to table TaxonNameProject |
| URI |
varchar (255) |
The complete URI address of the resource. |
NO |
- |
| Description |
xml (MAX) |
Description of the resource |
YES |
- |
| Notes |
nvarchar (MAX) |
A note concerning this entry. |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when the data were first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when the data were last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
| Title |
nvarchar (500) |
Title of the resource |
YES |
- |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
- |
| Creator |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
- |
| CreatorAgentURI |
varchar (500) |
Link to the module DiversityAgents |
YES |
- |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
- |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e. g. Creative Common Licenses |
YES |
- |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
- |
| LicenseHolderAgentURI |
varchar (500) |
The link to a module containing further information on the person or institution holding the license |
YES |
- |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
- |
| DisplayOrder |
int |
The sequence in which the resource should be displayed in a user interfaceDefault value: (1) |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise nullDefault value: '’ |
YES |
- |
| ResourceType |
nvarchar (50) |
Type of the resource as defined in table TaxonNameResourceType_Enum |
YES |
Refers to table TaxonNameResourceType_Enum |
Depending on:
- TaxonNameProject
- TaxonNameResourceType_Enum
Table TaxonNameTypification
The typification of a taxonomic name, e.g. where the type information of the name was published
| Column |
Data type |
Description |
Nullable |
Relation |
| NameID |
int |
The ID of a name in DiversityTaxonNames_Fungi (primary key) |
NO |
Refers to table TaxonName |
| TypificationReferenceTitle |
nvarchar (255) |
The title of the publication where the typification was published. Note this is only a cached value where Typification-LiteratureLink is present |
NO |
- |
| TypificationReferenceURI |
varchar (255) |
URI (e.g. LSID) of Reference, refers to e.g DiversityReferences: Source publication where the typification is published |
YES |
- |
| Typification |
nvarchar (500) |
The status of the type specimen(s) as written in the protologue, e.g. holotype |
YES |
- |
| TypificationDetails |
nvarchar (255) |
Details concerning the typification as written in the protologue, e.g. if just parts of a specimen were accepted as the type |
YES |
- |
| TypificationNotes |
nvarchar (200) |
Notes concerning the typification |
YES |
- |
| TypeSubstrate |
nvarchar (255) |
The substrate the type was growing on as written in the protologue |
YES |
- |
| TypeLocality |
nvarchar (255) |
The locality where the type was found as written in the protologue |
YES |
- |
| TypeSpecimenNotes |
nvarchar (500) |
Notes about the type specimen, e.g. the herbarium where a type specimen is located, Collector, collection date/number, etc. |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data. This is the operator (or typist) name, which may be different from the person responsible.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when record was first entered (typed or imported) into this system.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data. This is the operator (or typist) name, which may be different from the person responsible.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when record was last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
Table TaxonSynonymy
TaxonSynonymy contains names, which are synonym to other names. Here a single name may be a synonym of multiple accepted names (“pro parte synonym”). The “pro parte” or corresponding sensu information is captured in the “ConceptSuffix” attribute, which is part of the primary key.
| Column |
Data type |
Description |
Nullable |
Relation |
| ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module.Default value: (0) |
NO |
Refers to table ProjectProxy and table TaxonNameProject |
| NameID |
int |
ID of the synonymized name. Refers to the NameID of TaxonName (= foreign key). |
NO |
Refers to table TaxonNameProject |
| SynNameID |
int |
The ID of the synonym to which this name has been assigned to. Refers to the NameID of TaxonName (= foreign key). |
NO |
Refers to table TaxonNameProject |
| IgnoreButKeepForReference |
tinyint |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead.Default value: (0) |
NO |
- |
| ConceptSuffix |
nvarchar (200) |
Empty for default and primary concept; else ’s. lat.’, ’s. str.’, ‘sec.’, ‘sensu Muell.’, ’emend.’ , ‘(Auct.)’ etc.Default value: '’ |
YES |
- |
| ConceptNotes |
nvarchar (500) |
Notes on the concept of the name, e.g. incl. forms with black apothecia |
YES |
- |
| SynRefURI |
varchar (255) |
Reference URI: Source publication where synonymization is published (not publication of name!) as stored e.g. in DiversityReferences |
YES |
- |
| SynRefText |
nvarchar (255) |
Free text, esp. where a SynRefURI is missing. Source publication where synonymization is published (not publication of name!) |
YES |
- |
| SynRefDetail |
nvarchar (255) |
Esp. page number on which the synonymization is published |
YES |
- |
| SynTypistsNotes |
nvarchar (255) |
An internal note of the responsible person concerning this synonymization. This information is NOT included in any report. |
YES |
- |
| SynType |
nvarchar (50) |
Type of the synonymization, e.g. ‘heterotypic’, ‘homotypic’Default value: ‘unknown’ |
NO |
Refers to table TaxonNameSynonymisationType_Enum |
| SynIsUncertain |
bit |
If the synoymization is uncertain. Corresponding to =? for heterotypic synonyms and ? if the type of the synonymisation is unknown. Homotypic synonyms can not be uncertain.Default value: (0) |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data. This is the operator (or typist) name, which may be different from the person responsible.Default value: user_name() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when record was first entered (typed or imported) into this system.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data. This is the operator (or typist) name, which may be different from the person responsible.Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when record was last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
- ProjectProxy
- TaxonNameProject
- TaxonNameSynonymisationType_Enum
trgInsTaxonSynonymy
Setting version of taxon
VIEWS
View AcceptedNameChanges
Listing NameID of previous accepted names and the NameID of the current accepted name
| Column |
Data type |
Description |
Nullable |
| ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module. |
YES |
| NameID |
int |
ID of the accepted name. Refers to the NameID of TaxonName (= foreign key). |
YES |
| IgnoreButKeepForReference |
tinyint |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead. |
YES |
| LogDate |
datetime |
Date of the changes according to the log table |
NO |
| LogID |
int |
ID of the changes in the log table |
NO |
| CurrentAcceptedNameID |
int |
NameID of the accepted name |
YES |
Depending on:
- TaxonAcceptedName
- TaxonAcceptedName_log
- TaxonSynonymy
View GenusList
All Taxa with rank genus
| Column |
Data type |
Description |
Nullable |
| NameID |
int |
The ID of a name in DiversityTaxonNames_Fungi (primary key) |
NO |
| TaxonNameCache |
nvarchar (200) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
YES |
Depending on:
View NameID_UserAvailable
List of NameIDs accessible for the current user
| Column |
Data type |
Description |
Nullable |
| NameID |
int |
The ID of a name in DiversityTaxonNames (foreign key + part of primary key: the name may occur in multiple projects) |
NO |
Depending on:
- ProjectUser
- TaxonNameProject
View ProjectList
List of projects accessible for the current user
| Column |
Data type |
Description |
Nullable |
| Project |
nvarchar (50) |
The name of the project |
NO |
| ProjectID |
int |
ID of the project |
NO |
| ProjectURI |
varchar (255) |
The URI of the project, e.g. as provided by the module DiversityProjects. |
YES |
Depending on:
View TaxonAcceptedName_Core
Content of TaxonAcceptedName restricted to datasets accessible for a user
| Column |
Data type |
Description |
Nullable |
| ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module. |
NO |
| NameID |
int |
ID of the accepted name. Refers to the NameID of TaxonName (= foreign key). |
NO |
| IgnoreButKeepForReference |
tinyint |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead. |
NO |
| ConceptSuffix |
nvarchar (200) |
Empty for default and primary concept; else ’s. lat.’, ’s. str.’, ‘sec.’, ‘sensu Muell.’, ’emend.’ , ‘(Auct.)’ etc. |
YES |
| ConceptNotes |
nvarchar (500) |
Notes on the concept of the name, e.g. incl. forms with black apothecia |
YES |
| RefURI |
varchar (255) |
ReferenceURI: Source publication where synonymization is published (not publication of name!) |
YES |
| RefText |
nvarchar (255) |
Free text, esp. where a RefURI is missing. Source publication where synonymization is published (not publication of name!) |
YES |
| RefDetail |
nvarchar (255) |
Esp. page number on which the synonymization is published |
YES |
| TypistsNotes |
nvarchar (255) |
An internal note of the responsible person concerning this synonymization. This information is NOT included in any report. |
YES |
Depending on:
- NameID_UserAvailable
- TaxonAcceptedName
View TaxonAcceptedName_Indicated
List of IDs of accepted names
| Column |
Data type |
Description |
Nullable |
| ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module. |
NO |
| NameID |
int |
ID of the accepted name. Refers to the NameID of TaxonName (= foreign key). |
NO |
| IgnoreButKeepForReference |
tinyint |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead. |
NO |
| ConceptSuffix |
nvarchar (200) |
Empty for default and primary concept; else ’s. lat.’, ’s. str.’, ‘sec.’, ‘sensu Muell.’, ’emend.’ , ‘(Auct.)’ etc. |
YES |
| ConceptNotes |
nvarchar (500) |
Notes on the concept of the name, e.g. incl. forms with black apothecia |
YES |
| RefURI |
varchar (255) |
ReferenceURI: Source publication where synonymization is published (not publication of name!) |
YES |
| RefText |
nvarchar (255) |
Free text, esp. where a RefURI is missing. Source publication where synonymization is published (not publication of name!) |
YES |
| RefDetail |
nvarchar (255) |
Esp. page number on which the synonymization is published |
YES |
| TypistsNotes |
nvarchar (255) |
An internal note of the responsible person concerning this synonymization. This information is NOT included in any report. |
YES |
Depending on:
View TaxonAcceptedNameList
List of accepted names excluding ignored names
| Column |
Data type |
Description |
Nullable |
| NameID |
int |
ID of the accepted name. Refers to the NameID of TaxonName (= foreign key). |
NO |
| DisplayText |
nvarchar (4000) |
Combining taxon, publication and nomenclatural information |
YES |
| TaxonName |
nvarchar (301) |
Combining taxon information excluding authors |
YES |
| TaxonomicRank |
nvarchar (52) |
Taxonomic rank of the taxon (var., subsp., species, genus, family, order, etc.). The rank must be selected from the associated list of ranks (= TaxonomyRank) |
NO |
| InfraspecificEpithet |
nvarchar (101) |
The epithet of the infraspecific entity |
YES |
| Authors |
nvarchar (719) |
Combining all author information of the taxon |
YES |
| SynonymText |
nvarchar (1683) |
Combining publication and nomenclatural information |
YES |
| YearOfPubl |
smallint |
The year when the name was published |
YES |
| TaxonNameCache |
nvarchar (255) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
YES |
| ConceptSuffix |
nvarchar (200) |
Empty for default and primary concept; else ’s. lat.’, ’s. str.’, ‘sec.’, ‘sensu Muell.’, ’emend.’ , ‘(Auct.)’ etc. |
YES |
| ConceptNotes |
nvarchar (500) |
Notes on the concept of the name, e.g. incl. forms with black apothecia |
YES |
| BasedOnNameID |
int |
The Basionym of this name, resp. the NameID of the Basionym. A name is a Basionym, if NameID = BasionymID. |
YES |
| ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module. |
NO |
| NomenclaturalStatus |
nvarchar (50) |
Categories for effective/valid/legitimate… esp. ’nom. illeg.’, ’nom. inval.’, ’nom. nudum’, etc. |
YES |
Depending on:
- TaxonAcceptedName
- TaxonName
View TaxonAcceptedNameListIncludeIgnored
List of accepted names including ignored names
| Column |
Data type |
Description |
Nullable |
| NameID |
int |
ID of the accepted name. Refers to the NameID of TaxonName (= foreign key). |
NO |
| DisplayText |
nvarchar (2609) |
Combining taxon, publication and nomenclatural information |
YES |
| TaxonName |
nvarchar (301) |
Combining taxon information excluding authors |
YES |
| TaxonomicRank |
nvarchar (52) |
Taxonomic rank of the taxon (var., subsp., species, genus, family, order, etc.). The rank must be selected from the associated list of ranks (= TaxonomyRank) |
NO |
| InfraspecificEpithet |
nvarchar (101) |
The epithet of the infraspecific entity |
YES |
| Authors |
nvarchar (719) |
Combining all author information of the taxon |
YES |
| SynonymText |
nvarchar (1338) |
Combining publication and nomenclatural information |
YES |
| YearOfPubl |
smallint |
The year when the name was published |
YES |
| TaxonNameCache |
nvarchar (255) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
YES |
| ConceptSuffix |
nvarchar (200) |
Empty for default and primary concept; else ’s. lat.’, ’s. str.’, ‘sec.’, ‘sensu Muell.’, ’emend.’ , ‘(Auct.)’ etc. |
YES |
| ConceptNotes |
nvarchar (500) |
Notes on the concept of the name, e.g. incl. forms with black apothecia |
YES |
| BasedOnNameID |
int |
The Basionym of this name, resp. the NameID of the Basionym. A name is a Basionym, if NameID = BasionymID. |
YES |
| ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module. |
NO |
| NomenclaturalStatus |
nvarchar (50) |
Categories for effective/valid/legitimate… esp. ’nom. illeg.’, ’nom. inval.’, ’nom. nudum’, etc. |
YES |
| IgnoreButKeepForReference |
bit |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead. |
YES |
Depending on:
- TaxonAcceptedName
- TaxonName
View TaxonFamily
Family information for taxa
| Column |
Data type |
Description |
Nullable |
| NameID |
int |
Unique NameID code of the higher taxon. Refers to the NameID code of LichenName (= foreign key). |
NO |
| TaxonNameCache |
nvarchar (255) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
YES |
| FamilyNameID |
int |
NameID for the family |
YES |
| Family |
nvarchar (200) |
The name of the family |
YES |
| IgnoreButKeepForReference |
int |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead. |
NO |
Depending on:
- TaxonHierarchy
- TaxonName
- TaxonNameTaxonomicRank_Enum
View TaxonGeography_Core
User available information of the geography
| Column |
Data type |
Description |
Nullable |
| NameID |
int |
ID of the name. Refers to the NameID of TaxonName (= foreign key). |
NO |
| PlaceURI |
varchar (255) |
URI (e.g. LSID) within e.g. the DiversityGazetteer for a place (which may have several names) as found in the protologue |
NO |
| PlaceNameCache |
nvarchar (100) |
The name of the place |
YES |
Depending on:
- NameID_UserAvailable
- TaxonGeography
View TaxonHierarchy_Core
User available information of the hierarchy
| Column |
Data type |
Description |
Nullable |
| ProjectID |
int |
Each project can implement a different taxonomic hierarchy. Refers to the common project definition in the DiversityProjects module. |
NO |
| NameID |
int |
Unique NameID code of the higher taxon. Refers to the NameID code of LichenName (= foreign key). |
NO |
| IgnoreButKeepForReference |
tinyint |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead. |
NO |
| NameParentID |
int |
Next higher taxon (e.g. the family or subfamily if this taxon is a genus) |
YES |
| HierarchyRefURI |
varchar (255) |
Reference URI: Source publication where the hierarchy is published (not publication of name!) as stored e.g. in DiversityReferences |
YES |
| HierarchyRefText |
nvarchar (255) |
Free citation, esp. where a HierarchyRefURI is missing. Source publication where the hierarchy is published (not publication of name!) |
YES |
| HierarchyRefDetail |
nvarchar (255) |
Esp. page number on which the hierarchy is published |
YES |
| HierarchyPositionIsUncertain |
bit |
If the position of this taxon within this taxonomic group is uncertain. |
YES |
| HierarchyTypistsNotes |
nvarchar (255) |
An internal note of the responsible person concerning this hierarchy. This information is NOT included in any report. |
YES |
| HierarchyListCache |
nvarchar (1000) |
CALCULATED FIELD: List of higher taxa for faster access: “div.; class; ord.; fam.” |
YES |
Depending on:
- NameID_UserAvailable
- TaxonHierarchy
View TaxonListSynType
Public available information for taxa including synonymy
| Column |
Data type |
Description |
Nullable |
| NameID |
int |
ID of the accepted name. Refers to the NameID of TaxonName (= foreign key). |
NO |
| TaxonNameCache |
nvarchar (255) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
YES |
| BasedOnNameID |
int |
The Basionym of this name, resp. the NameID of the Basionym. A name is a Basionym, if NameID = BasionymID. |
YES |
| CreationType |
nvarchar (50) |
E.g.: taxon based on new type, combination based on a previously publ. name (‘comb. nov.’), new name (’nom.nov.’) introduced to replace a homonym (may occur for genera!), validation of previously invalidly publ. taxon name (’ex’), or unknown. |
YES |
| TaxonomicRank |
nvarchar (50) |
Taxonomic rank of the taxon (var., subsp., species, genus, family, order, etc.). The rank must be selected from the associated list of ranks (= TaxonomyRank) |
NO |
| GenusOrSupragenericName |
nvarchar (200) |
If rank is above species: Name of taxon above species level (currently accepted spelling). Includes infrageneric taxon names, genera, families, etc. |
NO |
| SpeciesGenusNameID |
int |
If rank is species or below: NameID of the Genus name. Refers to same table with TaxonomicRank = genus. |
YES |
| InfragenericEpithet |
nvarchar (200) |
If rank below genus and above species: Name of infrageneric taxon above species level (currently accepted spelling). Includes subgenus, series, etc. |
YES |
| SpeciesEpithet |
nvarchar (100) |
The species name part of the species name, for example ‘alba’ in ‘Abies alba’. |
YES |
| InfraspecificEpithet |
nvarchar (100) |
The epithet of the infraspecific entity |
YES |
| BasionymAuthors |
nvarchar (100) |
The authors of a newly created name. For all taxonomic ranks, only for ‘comb. nov.’ or ’nom. nov.’: Author(s) of the basionym (will be displayed in ‘()’, do not enter the parentheses), abbreviated according to authors standard |
YES |
| CombiningAuthors |
nvarchar (255) |
The names of the combining authors if the name is base on another older name (e.g. combined into a different genus) |
YES |
| PublishingAuthors |
nvarchar (255) |
If the authors of the taxon differ from the authors of the publication: the latter (…‘in ’ Publishing authors … ) |
YES |
| SanctioningAuthor |
nvarchar (100) |
Sanctioning is a special instrument available for fungi to allow the starting date of taxonomy to be identical with higher plants, but preserve names used by Fries (Fr.) and Person (Pers.) |
YES |
| NonNomenclaturalNameSuffix |
nvarchar (200) |
A suffix for concept names not included in any nomenclatural code. If present fields for authors and typification must be empty and NomenclaturalCode set to ’not under code’. |
YES |
| IsHybrid |
bit |
If checked (or ‘True’), the new taxon is a hybrid with or without a hybrid epithet. |
YES |
| HybridNameID1 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 1. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
| HybridNameID2 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 2. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
| HybridNameID3 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 3. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
| HybridNameID4 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 4. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
| ReferenceTitle |
nvarchar (600) |
The title of the publication where the name was published. Note this is only a cached value where ReferenceURI is present |
YES |
| ReferenceURI |
varchar (255) |
URI (e.g. LSID) of Reference, referes to table ReferenceTitle in Database DiversityReferences: Source publication where name is published |
YES |
| Volume |
nvarchar (20) |
The volume of the journal |
YES |
| Issue |
nvarchar (255) |
The issue of the literature |
YES |
| Pages |
nvarchar (50) |
The pages within the literature |
YES |
| YearOfPubl |
smallint |
The year when the name was published |
YES |
| YearOnPubl |
smallint |
The year cited on the original paper as year of publication |
YES |
| NameUsageReferences |
nvarchar (255) |
Indexing volumes like Index of Fungi or any Name usage that caused this name to be added to the database |
|
| YES |
|
|
|
| OriginalOrthography |
nvarchar (255) |
The original spelling of the name |
YES |
| NomenclaturalCode |
nvarchar (50) |
Code of Nomenclature under which this taxon was created: ‘Bacteriology’, ‘Botany’ (incl. Mycology), ‘Zoology’, ‘Biocode’ (for future use), ‘Non nomenclatural name’. Default value: ‘Botany’ |
YES |
| NomenclaturalStatus |
nvarchar (50) |
Categories for effective/valid/legitimate… esp. ’nom. illeg.’, ’nom. inval.’, ’nom. nudum’, etc. |
YES |
| NomenclaturalComment |
nvarchar (MAX) |
Comments on the nomenclature. e.g. ‘according to ICBN Art. 39.1’ |
YES |
| Typification |
nvarchar (50) |
The status of the type specimen(s) as written in the protologue, e.g. holotype |
YES |
| TypificationDetails |
nvarchar (255) |
Details concerning the typification as written in the protologue, e.g. if just parts of a specimen were accepted as the type |
YES |
| TypeSubstrate |
nvarchar (255) |
The substrate the type was growing on as written in the protologue |
YES |
| TypeLocality |
nvarchar (255) |
The locality where the type was found as written in the protologue |
YES |
| TypificationReferenceTitle |
nvarchar (255) |
The title of the publication where the typification was published. Note this is only a cached value where Typification-LiteratureLink is present |
YES |
| TypificationReferenceURI |
varchar (255) |
URI (e.g. LSID) of Reference, refers to e.g DiversityReferences: Source publication where the typification is published |
YES |
| TypificationNotes |
nvarchar (200) |
Notes concerning the typification |
YES |
| AnamorphTeleomorph |
nvarchar (255) |
If the name is related to a anamorph or a teleomorph as written in the protologue |
YES |
| TypistNotes |
nvarchar (200) |
Additional notes and problems |
YES |
| SynType |
nvarchar (50) |
Type of the synonymization, e.g. ‘heterotypic’, ‘homotypic’ |
YES |
| ValidSynType |
nvarchar (50) |
“accepted name” or type of the synonymy |
YES |
| IgnoreAcceptedName |
int |
If accepted name is ignored (1 = ignored, 0 = not ignored) |
NO |
| IgnoreSynonymy |
int |
If synonymy is ignored (1 = ignored, 0 = not ignored) |
NO |
Depending on:
- TaxonAcceptedName
- TaxonName
- TaxonSynonymy
View TaxonName_Core
User available taxa
| Column |
Data type |
Description |
Nullable |
| NameID |
int |
The ID of a name in DiversityTaxonNames_Fungi (primary key) |
NO |
| Display |
nvarchar (255) |
Corresponds to TaxonNameCache |
YES |
| DisplayEpithet |
nvarchar (358) |
SpeciesEpithet including information for whole taxon for query |
YES |
| TaxonNameCache |
nvarchar (255) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
YES |
| Version |
smallint |
The version of a name record (revision number, internally filled by system) |
YES |
| BasedOnNameID |
int |
The Basionym of this name, resp. the NameID of the Basionym. A name is a Basionym, if NameID = BasionymID. |
YES |
| CreationType |
nvarchar (50) |
E.g.: taxon based on new type, combination based on a previously publ. name (‘comb. nov.’), new name (’nom.nov.’) introduced to replace a homonym (may occur for genera!), validation of previously invalidly publ. taxon name (’ex’), or unknown. |
YES |
| TaxonomicRank |
nvarchar (50) |
Taxonomic rank of the taxon (var., subsp., species, genus, family, order, etc.). The rank must be selected from the associated list of ranks (= TaxonomyRank) |
NO |
| GenusOrSupragenericName |
nvarchar (200) |
If rank is above species: Name of taxon above species level (currently accepted spelling). Includes infrageneric taxon names, genera, families, etc. |
NO |
| SpeciesGenusNameID |
int |
If rank is species or below: NameID of the Genus name. Refers to same table with TaxonomicRank = genus. |
YES |
| InfragenericEpithet |
nvarchar (200) |
If rank below genus and above species: Name of infrageneric taxon above species level (currently accepted spelling). Includes subgenus, series, etc. |
YES |
| SpeciesEpithet |
nvarchar (100) |
The species name part of the species name, for example ‘alba’ in ‘Abies alba’. |
YES |
| InfraspecificEpithet |
nvarchar (100) |
The epithet of the infraspecific entity |
YES |
| BasionymAuthors |
nvarchar (100) |
The authors of a newly created name. For all taxonomic ranks, only for ‘comb. nov.’ or ’nom. nov.’: Author(s) of the basionym (will be displayed in ‘()’, do not enter the parentheses), abbreviated according to authors standard |
YES |
| CombiningAuthors |
nvarchar (255) |
The names of the combining authors if the name is base on another older name (e.g. combined into a different genus) |
YES |
| PublishingAuthors |
nvarchar (255) |
If the authors of the taxon differ from the authors of the publication: the latter (…‘in ’ Publishing authors … ) |
YES |
| SanctioningAuthor |
nvarchar (100) |
Sanctioning is a special instrument available for fungi to allow the starting date of taxonomy to be identical with higher plants, but preserve names used by Fries (Fr.) and Person (Pers.) |
YES |
| NonNomenclaturalNameSuffix |
nvarchar (200) |
A suffix for concept names not included in any nomenclatural code. If present fields for authors and typification must be empty and NomenclaturalCode set to ’not under code’. |
YES |
| IsHybrid |
bit |
If checked (or ‘True’), the new taxon is a hybrid with or without a hybrid epithet. |
YES |
| HybridNameID1 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 1. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
| HybridNameID2 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 2. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
| HybridNameID3 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 3. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
| HybridNameID4 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 4. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
| ReferenceTitle |
nvarchar (600) |
The title of the publication where the name was published. Note this is only a cached value where ReferenceURI is present |
YES |
| ReferenceURI |
varchar (255) |
URI (e.g. LSID) of Reference, referes to table ReferenceTitle in Database DiversityReferences: Source publication where name is published |
YES |
| Volume |
nvarchar (20) |
The volume of the journal |
YES |
| Issue |
nvarchar (255) |
The issue of the literature |
YES |
| Pages |
nvarchar (50) |
The pages within the literature |
YES |
| DetailLocation |
nvarchar (200) |
Additional information like plates etc. |
YES |
| DayOfPubl |
tinyint |
The day when the name was published |
YES |
| MonthOfPubl |
tinyint |
The month when the name was published |
YES |
| YearOfPubl |
smallint |
The year when the name was published |
YES |
| DateOfPublSupplement |
nvarchar (200) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
YES |
| YearOnPubl |
smallint |
The year cited on the original paper as year of publication |
YES |
| DateOnPublSupplement |
nvarchar (200) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
YES |
| Protologue |
nvarchar (MAX) |
Full text of the protologue. If protologue is provided in Latin and in another language |
|
| languages, both only the Latin or several languages may be stored together. |
YES |
|
|
| ProtologueURI |
varchar (255) |
URI of externally available Protologue information (scanned image, full text from external provider, etc.) |
YES |
| ProtologueResourceURI |
varchar (255) |
The ResourceID of an image of the protologue as stored in the module DiversityResources. Primary key of table Resource in the database DiversityResources.(= Foreign key) |
YES |
| NameUsageReferences |
nvarchar (255) |
Indexing volumes like Index of Fungi or any Name usage that caused this name to be added to the database |
|
| YES |
|
|
|
| OriginalOrthography |
nvarchar (255) |
The original spelling of the name |
YES |
| NomenclaturalCode |
nvarchar (50) |
Code of Nomenclature under which this taxon was created: ‘Bacteriology’, ‘Botany’ (incl. Mycology), ‘Zoology’, ‘Biocode’ (for future use), ‘Non nomenclatural name’. Default value: ‘Botany’ |
YES |
| NomenclaturalStatus |
nvarchar (50) |
Categories for effective/valid/legitimate… esp. ’nom. illeg.’, ’nom. inval.’, ’nom. nudum’, etc. |
YES |
| NomenclaturalComment |
nvarchar (MAX) |
Comments on the nomenclature. e.g. ‘according to ICBN Art. 39.1’ |
YES |
| Typification |
nvarchar (50) |
The status of the type specimen(s) as written in the protologue, e.g. holotype |
YES |
| TypificationDetails |
nvarchar (255) |
Details concerning the typification as written in the protologue, e.g. if just parts of a specimen were accepted as the type |
YES |
| TypificationReferenceTitle |
nvarchar (255) |
The title of the publication where the typification was published. Note this is only a cached value where Typification-LiteratureLink is present |
YES |
| TypificationReferenceURI |
varchar (255) |
URI (e.g. LSID) of Reference, refers to e.g DiversityReferences: Source publication where the typification is published |
YES |
| TypificationNotes |
nvarchar (200) |
Notes concerning the typification |
YES |
| TypeSubstrate |
nvarchar (255) |
The substrate the type was growing on as written in the protologue |
YES |
| TypeLocality |
nvarchar (255) |
The locality where the type was found as written in the protologue |
YES |
| TypeSpecimenNotes |
nvarchar (200) |
Notes about the type specimen, e.g. the herbarium where a type specimen is located, Collector, collection date/number, etc. |
YES |
| AnamorphTeleomorph |
nvarchar (255) |
If the name is related to a anamorph or a teleomorph as written in the protologue |
YES |
| TypistNotes |
nvarchar (200) |
Additional notes and problems |
YES |
| RevisionLevel |
nvarchar (50) |
The level of the revision of the taxonomic name, e.g. ‘unchecked’, ‘fully checked’ |
YES |
| IgnoreButKeepForReference |
bit |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead. |
YES |
Depending on:
- NameID_UserAvailable
- TaxonName
View TaxonName_Indicated
Content of table TaxonName including synonymy information
| Column |
Data type |
Description |
Nullable |
| ProjectID |
int |
ID of the project. Refers to the common project definition in the DiversityProjects module. |
NO |
| Display |
nvarchar (263) |
Content of column TaxonNameCache with synonymy prefix |
YES |
| DisplayEpithet |
nvarchar (366) |
Content of column SpeciesEpithet with synonymy prefix and trailed by TaxonNameCache |
YES |
| NameID |
int |
The ID of a name in DiversityTaxonNames_Fungi (primary key) |
NO |
| TaxonNameCache |
nvarchar (255) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
YES |
| Version |
smallint |
The version of a name record (revision number, internally filled by system) |
YES |
| BasedOnNameID |
int |
The Basionym of this name, resp. the NameID of the Basionym. A name is a Basionym, if NameID = BasionymID. |
YES |
| CreationType |
nvarchar (50) |
E.g.: taxon based on new type, combination based on a previously publ. name (‘comb. nov.’), new name (’nom.nov.’) introduced to replace a homonym (may occur for genera!), validation of previously invalidly publ. taxon name (’ex’), or unknown. |
YES |
| TaxonomicRank |
nvarchar (50) |
Taxonomic rank of the taxon (var., subsp., species, genus, family, order, etc.). The rank must be selected from the associated list of ranks (= TaxonomyRank) |
NO |
| GenusOrSupragenericName |
nvarchar (200) |
If rank is above species: Name of taxon above species level (currently accepted spelling). Includes infrageneric taxon names, genera, families, etc. |
NO |
| SpeciesGenusNameID |
int |
If rank is species or below: NameID of the Genus name. Refers to same table with TaxonomicRank = genus. |
YES |
| InfragenericEpithet |
nvarchar (200) |
If rank below genus and above species: Name of infrageneric taxon above species level (currently accepted spelling). Includes subgenus, series, etc. |
YES |
| SpeciesEpithet |
nvarchar (100) |
The species name part of the species name, for example ‘alba’ in ‘Abies alba’. |
YES |
| InfraspecificEpithet |
nvarchar (100) |
The epithet of the infraspecific entity |
YES |
| BasionymAuthors |
nvarchar (100) |
The authors of a newly created name. For all taxonomic ranks, only for ‘comb. nov.’ or ’nom. nov.’: Author(s) of the basionym (will be displayed in ‘()’, do not enter the parentheses), abbreviated according to authors standard |
YES |
| CombiningAuthors |
nvarchar (255) |
The names of the combining authors if the name is base on another older name (e.g. combined into a different genus) |
YES |
| PublishingAuthors |
nvarchar (255) |
If the authors of the taxon differ from the authors of the publication: the latter (…‘in ’ Publishing authors … ) |
YES |
| SanctioningAuthor |
nvarchar (100) |
Sanctioning is a special instrument available for fungi to allow the starting date of taxonomy to be identical with higher plants, but preserve names used by Fries (Fr.) and Person (Pers.) |
YES |
| NonNomenclaturalNameSuffix |
nvarchar (200) |
A suffix for concept names not included in any nomenclatural code. If present fields for authors and typification must be empty and NomenclaturalCode set to ’not under code’. |
YES |
| IsHybrid |
bit |
If checked (or ‘True’), the new taxon is a hybrid with or without a hybrid epithet. |
YES |
| HybridNameID1 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 1. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
| HybridNameID2 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 2. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
| HybridNameID3 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 3. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
| HybridNameID4 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 4. Refers to NomID code of publ. TaxonomyName (= foreign key). |
YES |
| ReferenceTitle |
nvarchar (600) |
The title of the publication where the name was published. Note this is only a cached value where ReferenceURI is present |
YES |
| ReferenceURI |
varchar (255) |
URI (e.g. LSID) of Reference, referes to table ReferenceTitle in Database DiversityReferences: Source publication where name is published |
YES |
| Volume |
nvarchar (20) |
The volume of the journal |
YES |
| Issue |
nvarchar (255) |
The issue of the literature |
YES |
| Pages |
nvarchar (50) |
The pages within the literature |
YES |
| DetailLocation |
nvarchar (200) |
Additional information like plates etc. |
YES |
| DayOfPubl |
tinyint |
The day when the name was published |
YES |
| MonthOfPubl |
tinyint |
The month when the name was published |
YES |
| YearOfPubl |
smallint |
The year when the name was published |
YES |
| DateOfPublSupplement |
nvarchar (200) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
YES |
| YearOnPubl |
smallint |
The year cited on the original paper as year of publication |
YES |
| DateOnPublSupplement |
nvarchar (200) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
YES |
| Protologue |
nvarchar (MAX) |
Full text of the protologue. If protologue is provided in Latin and in another language |
|
| languages, both only the Latin or several languages may be stored together. |
YES |
|
|
| ProtologueURI |
varchar (255) |
URI of externally available Protologue information (scanned image, full text from external provider, etc.) |
YES |
| ProtologueResourceURI |
varchar (255) |
The ResourceID of an image of the protologue as stored in the module DiversityResources. Primary key of table Resource in the database DiversityResources.(= Foreign key) |
YES |
| NameUsageReferences |
nvarchar (255) |
Indexing volumes like Index of Fungi or any Name usage that caused this name to be added to the database |
|
| YES |
|
|
|
| OriginalOrthography |
nvarchar (255) |
The original spelling of the name |
YES |
| NomenclaturalCode |
nvarchar (50) |
Code of Nomenclature under which this taxon was created: ‘Bacteriology’, ‘Botany’ (incl. Mycology), ‘Zoology’, ‘Biocode’ (for future use), ‘Non nomenclatural name’. Default value: ‘Botany’ |
YES |
| NomenclaturalStatus |
nvarchar (50) |
Categories for effective/valid/legitimate… esp. ’nom. illeg.’, ’nom. inval.’, ’nom. nudum’, etc. |
YES |
| NomenclaturalComment |
nvarchar (MAX) |
Comments on the nomenclature. e.g. ‘according to ICBN Art. 39.1’ |
YES |
| Typification |
nvarchar (50) |
The status of the type specimen(s) as written in the protologue, e.g. holotype |
YES |
| TypificationDetails |
nvarchar (255) |
Details concerning the typification as written in the protologue, e.g. if just parts of a specimen were accepted as the type |
YES |
| TypificationReferenceTitle |
nvarchar (255) |
The title of the publication where the typification was published. Note this is only a cached value where Typification-LiteratureLink is present |
YES |
| TypificationReferenceURI |
varchar (255) |
URI (e.g. LSID) of Reference, refers to e.g DiversityReferences: Source publication where the typification is published |
YES |
| TypificationNotes |
nvarchar (200) |
Notes concerning the typification |
YES |
| TypeSubstrate |
nvarchar (255) |
The substrate the type was growing on as written in the protologue |
YES |
| TypeLocality |
nvarchar (255) |
The locality where the type was found as written in the protologue |
YES |
| TypeSpecimenNotes |
nvarchar (200) |
Notes about the type specimen, e.g. the herbarium where a type specimen is located, Collector, collection date/number, etc. |
YES |
| AnamorphTeleomorph |
nvarchar (255) |
If the name is related to a anamorph or a teleomorph as written in the protologue |
YES |
| TypistNotes |
nvarchar (200) |
Additional notes and problems |
YES |
| RevisionLevel |
nvarchar (50) |
The level of the revision of the taxonomic name, e.g. ‘unchecked’, ‘fully checked’ |
YES |
| IgnoreButKeepForReference |
bit |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead. |
YES |
Depending on:
- ProjectList
- TaxonAcceptedName_Indicated
- TaxonName
- TaxonNameProject
- TaxonSynonymy_Indicated
View TaxonNameExternalID_Core
User available content of table TaxonNameExternalID
| Column |
Data type |
Description |
Nullable |
| NameID |
int |
The ID of a name in DiversityTaxonNames (foreign key + part of primary key: the name string associated with NameID may occur in multiple external databases) |
NO |
| ExternalDatabaseID |
int |
The ID of an external taxonomic data collection as defined in TaxontNameExternalDatabase (foreign key + part of primary key) |
NO |
| ExternalNameURI |
varchar (255) |
The URI (e.g. a LSID) of the external name as defined in the external database |
YES |
Depending on:
- NameID_UserAvailable
- TaxonNameExternalID
View TaxonOrder
Not ignored Order for taxa
| Column |
Data type |
Description |
Nullable |
| NameID |
int |
Unique NameID code of the higher taxon. Refers to the NameID code of LichenName (= foreign key). |
NO |
| TaxonNameCache |
nvarchar (255) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
YES |
| OrderNameID |
int |
The NameID of the Order |
YES |
| Order |
nvarchar (200) |
The Name of the Order |
YES |
| IgnoreButKeepForReference |
int |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead. |
NO |
Depending on:
View TaxonSynonymy_Core
User available content of table TaxonSynonymy
| Column |
Data type |
Description |
Nullable |
| ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module. |
NO |
| NameID |
int |
ID of the synonymized name. Refers to the NameID of TaxonName (= foreign key). |
NO |
| SynNameID |
int |
The ID of the synonym to which this name has been assigned to. Refers to the NameID of TaxonName (= foreign key). |
NO |
| IgnoreButKeepForReference |
tinyint |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead. |
NO |
| ConceptSuffix |
nvarchar (200) |
Empty for default and primary concept; else ’s. lat.’, ’s. str.’, ‘sec.’, ‘sensu Muell.’, ’emend.’ , ‘(Auct.)’ etc. |
NO |
| ConceptNotes |
nvarchar (500) |
Notes on the concept of the name, e.g. incl. forms with black apothecia |
YES |
| SynRefURI |
varchar (255) |
Reference URI: Source publication where synonymization is published (not publication of name!) as stored e.g. in DiversityReferences |
YES |
| SynRefText |
nvarchar (255) |
Free text, esp. where a SynRefURI is missing. Source publication where synonymization is published (not publication of name!) |
YES |
| SynRefDetail |
nvarchar (255) |
Esp. page number on which the synonymization is published |
YES |
| SynTypistsNotes |
nvarchar (255) |
An internal note of the responsible person concerning this synonymization. This information is NOT included in any report. |
YES |
| SynType |
nvarchar (50) |
Type of the synonymization, e.g. ‘heterotypic’, ‘homotypic’ |
YES |
| SynIsUncertain |
bit |
If the synoymization is uncertain. Corresponding to =? for heterotypic synonyms and ? if the type of the synonymisation is unknown. Homotypic synonyms can not be uncertain. |
YES |
Depending on:
- NameID_UserAvailable
- TaxonSynonymy
View TaxonSynonymy_Indicated
Prefix information for Synonyms
| Column |
Data type |
Description |
Nullable |
| ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module. |
NO |
| NameID |
int |
ID of the synonymized name. Refers to the NameID of TaxonName (= foreign key). |
NO |
| SynType |
nvarchar (50) |
Type of the synonymization, e.g. ‘heterotypic’, ‘homotypic’ |
YES |
| Prefix |
nvarchar (4) |
Prefix for Synonyms as displayed in query |
YES |
| Indent |
varchar (4) |
Indent for Synonyms as displayed in query |
YES |
Depending on:
View TaxonSynonymyAcceptedNameID
Accepted names for synonyms
| Column |
Data type |
Description |
Nullable |
| ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module. |
NO |
| NameID |
int |
ID of the accepted name. Refers to the NameID of TaxonName (= foreign key). |
NO |
| AcceptedNameID |
int |
ID of the accepted name |
YES |
Depending on:
- TaxonAcceptedName
- TaxonSynonymy
View TaxonSynonymyList
Taxon information including synonymy for not ignored data
| Column |
Data type |
Description |
Nullable |
| NameID |
int |
The ID of a name in DiversityTaxonNames_Fungi (primary key) |
NO |
| DisplayText |
nvarchar (4000) |
Taxon name including publication and nomenclatural information |
YES |
| TaxonName |
nvarchar (301) |
Taxonomic name excluding authors |
YES |
| TaxonomicRank |
nvarchar (52) |
Taxonomic rank of the taxon (var., subsp., species, genus, family, order, etc.). The rank must be selected from the associated list of ranks (= TaxonomyRank) |
NO |
| InfraspecificEpithet |
nvarchar (101) |
The epithet of the infraspecific entity |
YES |
| Authors |
nvarchar (922) |
Combining all author information of the taxon |
YES |
| SynonymText |
nvarchar (1683) |
Combining publication and nomenclatural information |
YES |
| TaxonNameCache |
nvarchar (255) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
YES |
| YearOfPubl |
smallint |
The year when the name was published |
YES |
| SynIsUncertain |
bit |
If the synoymization is uncertain. Corresponding to =? for heterotypic synonyms and ? if the type of the synonymisation is unknown. Homotypic synonyms can not be uncertain. |
YES |
| BasedOnNameID |
int |
The Basionym of this name, resp. the NameID of the Basionym. A name is a Basionym, if NameID = BasionymID. |
YES |
| ConceptSuffix |
nvarchar (200) |
Empty for default and primary concept; else ’s. lat.’, ’s. str.’, ‘sec.’, ‘sensu Muell.’, ’emend.’ , ‘(Auct.)’ etc. |
YES |
| ConceptNotes |
nvarchar (500) |
Notes on the concept of the name, e.g. incl. forms with black apothecia |
YES |
| SynType |
nvarchar (50) |
Type of the synonymization, e.g. ‘heterotypic’, ‘homotypic’ |
NO |
| SynNameID |
int |
The ID of the synonym to which this name has been assigned to. Refers to the NameID of TaxonName (= foreign key). |
NO |
| ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module. |
NO |
| NomenclaturalStatus |
nvarchar (50) |
Categories for effective/valid/legitimate… esp. ’nom. illeg.’, ’nom. inval.’, ’nom. nudum’, etc. |
YES |
Depending on:
View TaxonSynonymyListIncludeIgnored
Taxon information including synonymy and including ignored data
| Column |
Data type |
Description |
Nullable |
| NameID |
int |
The ID of a name in DiversityTaxonNames_Fungi (primary key) |
NO |
| DisplayText |
nvarchar (2613) |
Taxon name including publication and nomenclatural information |
YES |
| TaxonName |
nvarchar (301) |
Taxonomic name excluding authors |
YES |
| TaxonomicRank |
nvarchar (52) |
Taxonomic rank of the taxon (var., subsp., species, genus, family, order, etc.). The rank must be selected from the associated list of ranks (= TaxonomyRank) |
NO |
| InfraspecificEpithet |
nvarchar (101) |
The epithet of the infraspecific entity |
YES |
| Authors |
nvarchar (922) |
Combining all author information of the taxon |
YES |
| SynonymText |
nvarchar (1338) |
Combining publication and nomenclatural information |
YES |
| TaxonNameCache |
nvarchar (255) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
YES |
| YearOfPubl |
smallint |
The year when the name was published |
YES |
| SynIsUncertain |
bit |
If the synoymization is uncertain. Corresponding to =? for heterotypic synonyms and ? if the type of the synonymisation is unknown. Homotypic synonyms can not be uncertain. |
YES |
| BasedOnNameID |
int |
The Basionym of this name, resp. the NameID of the Basionym. A name is a Basionym, if NameID = BasionymID. |
YES |
| ConceptSuffix |
nvarchar (200) |
Empty for default and primary concept; else ’s. lat.’, ’s. str.’, ‘sec.’, ‘sensu Muell.’, ’emend.’ , ‘(Auct.)’ etc. |
YES |
| ConceptNotes |
nvarchar (500) |
Notes on the concept of the name, e.g. incl. forms with black apothecia |
YES |
| SynType |
nvarchar (50) |
Type of the synonymization, e.g. ‘heterotypic’, ‘homotypic’ |
NO |
| SynNameID |
int |
The ID of the synonym to which this name has been assigned to. Refers to the NameID of TaxonName (= foreign key). |
NO |
| ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module. |
NO |
| NomenclaturalStatus |
nvarchar (50) |
Categories for effective/valid/legitimate… esp. ’nom. illeg.’, ’nom. inval.’, ’nom. nudum’, etc. |
YES |
| IgnoreButKeepForReference |
bit |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead. |
YES |
Depending on:
View ViewBaseURL
Result of function dbo.BaseURL() for linked servers
| Column |
Data type |
Description |
Nullable |
| BaseURL |
varchar (255) |
The BaseURL of the database |
YES |
Depending on:
View ViewDefaultProjectID
Result of function dbo.DefaultProjectID() for linked servers
| Column |
Data type |
Description |
Nullable |
| ProjectID |
int |
ID of the project. Foreign key, relates to PK of table ProjectProxy |
YES |
Depending on:
View ViewDiversityWorkbenchModule
Result of function dbo.DiversityWorkbenchModule() for linked servers
| Column |
Data type |
Description |
Nullable |
| DiversityWorkbenchModule |
nvarchar (50) |
The DiversityWorkbenchModule of the database = DiversityTaxonNames |
YES |
Depending on:
View ViewTaxonNameResource
A View for TaxonNameResource excluding XML columns
| Column |
Data type |
Description |
Nullable |
| NameID |
int |
ID of the name. Refers to the NameID of TaxonName (= foreign key). |
NO |
| ProjectID |
int |
ID of the project. Refers to the common project definition in the DiversityProjects module. |
NO |
| URI |
varchar (255) |
The complete URI address of the resource. |
NO |
| Notes |
nvarchar (MAX) |
A note concerning this entry. |
YES |
| Title |
nvarchar (500) |
Title of the resource |
YES |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
| Creator |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
| CreatorAgentURI |
varchar (500) |
Link to the module DiversityAgents |
YES |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e. g. Creative Common Licenses |
YES |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
| LicenseHolderAgentURI |
varchar (500) |
The link to a module containing further information on the person or institution holding the license |
YES |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
| DisplayOrder |
int |
The sequence in which the resource should be displayed in a user interface |
YES |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
| ResourceType |
nvarchar (50) |
Type of the resource as defined in table TaxonNameResourceType_Enum |
YES |
Depending on:
FUNCTIONS
Function AcceptedNameID
Returns ID of accepted name if any is found, otherwise NULL
DataType: int
| Parameter |
DataType |
Description |
| @NameID |
int |
ID of the accepted name. Refers to the NameID of TaxonName (= foreign key). |
| @ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module. |
Depending on:
- DefaultProjectID
- TaxonAcceptedName
- TaxonName
- TaxonSynonymy
Function AnalysisCategoryHierarchyAll
Returns a table that lists all the analysis categorys.
| Column |
DataType |
Description |
| AnalysisID |
int |
ID of the analysis (Primary key) |
| AnalysisParentID |
int |
Analysis ID of the parent analysis if it belongs to a certain type documented in this table |
| DisplayText |
nvarchar (50) |
Name of the analysis as e.g. shown in user interface |
| Description |
nvarchar (MAX) |
Description of the analysis |
| Notes |
nvarchar (MAX) |
Notes concerning this analysis |
| AnalysisURI |
varchar (255) |
URI referring to an external documentation of the analysis |
| OnlyHierarchy |
bit |
Reserved for future application |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
| SortingID |
int |
An ID for sorting the data |
| HierarchyDisplayText |
varchar (900) |
DisplayTexts of the hierarchy separated by " |
Depending on:
- TaxonNameListAnalysisCategory
Function BaseURL
Returns BaseURL for the database
DataType: varchar (255)
Function DefaultProjectID
ID of the current project selected by the user
DataType: int
Depending on:
Function DiversityMobile_TaxonList
Returns a result set that lists all taxa in a given project resp. list
| Parameter |
DataType |
Description |
| @ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module. |
| @ListID |
int |
The ID of the List corresponding to column ProjectID in table TaxonNameList |
| Column |
DataType |
Description |
| NameURI |
varchar (255) |
Combines content of dbo.BaseURL() and NameID of the taxon |
| TaxonNameCache |
nvarchar (255) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
| TaxonNameSinAuthors |
nvarchar (255) |
Taxonomic name excluding authors |
| GenusOrSupragenericName |
nvarchar (255) |
If rank is above species: Name of taxon above species level (currently accepted spelling). Includes infrageneric taxon names, genera, families, etc. |
| SpeciesEpithet |
nvarchar (255) |
The species name part of the species name, for example ‘alba’ in ‘Abies alba’. |
| InfraspecificEpithet |
nvarchar (255) |
The epithet of the infraspecific entity |
| Synonymy |
varchar (50) |
Simple synonymy information for the taxon as either “Accepted” or “Synonym” |
| Family |
nvarchar (255) |
Family of the taxon |
| Order |
nvarchar (255) |
Order of the taxon |
Depending on:
- BaseURL
- TaxonAcceptedName
- TaxonHierarchy
- TaxonName
- TaxonNameList
- TaxonNameProject
- TaxonSynonymy
Function DiversityWorkbenchModule
Returns the DiversityWorkbenchModule = “DiversityTaxonNames”
DataType: nvarchar (50)
Function HierarchyCache
Returns a string of all the taxa superior to a given taxon separated with the value of parameter @Separator
DataType: nvarchar (-1)
| Parameter |
DataType |
Description |
| @NameID |
int |
Unique NameID code of the higher taxon. Refers to the NameID code of LichenName (= foreign key). |
| @DiversityProjectID |
int |
ID of the project |
| @Separator |
char (1) |
Separator for the hierarchy levels |
Depending on:
- ProjectList
- TaxonHierarchy
- TaxonHierarchy_Core
- TaxonName
- TaxonNameProject
Function HierarchyFamilyList
Returns a result set that lists all families of the taxa in a given project
| Parameter |
DataType |
Description |
| @ProjectID |
int |
Each project can implement a different taxonomic hierarchy. Refers to the common project definition in the DiversityProjects module. |
| Column |
DataType |
Description |
| NameID |
int |
Unique NameID code of the higher taxon. Refers to the NameID code of LichenName (= foreign key). |
| TaxonNameCache |
nvarchar (500) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
| FamilyNameID |
int |
NameID of the family |
| Family |
nvarchar (500) |
The name of the family |
Depending on:
Function HierarchyList
Returns a result set that lists hierarchy of the taxa in a given project
| Parameter |
DataType |
Description |
| @ProjectID |
int |
Each project can implement a different taxonomic hierarchy. Refers to the common project definition in the DiversityProjects module. |
| Column |
DataType |
Description |
| NameID |
int |
Unique NameID code of the higher taxon. Refers to the NameID code of LichenName (= foreign key). |
| TaxonNameCache |
nvarchar (500) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
| Hierarchy |
nvarchar (2000) |
Hierarchy above the taxon separated by " |
Depending on:
- TaxonHierarchy
- TaxonName
- TaxonNameTaxonomicRank_Enum
Function HierarchyOrderList
Returns a result set that lists all orders of the taxa in a given project.
| Parameter |
DataType |
Description |
| @ProjectID |
int |
Each project can implement a different taxonomic hierarchy. Refers to the common project definition in the DiversityProjects module. |
| Column |
DataType |
Description |
| NameID |
int |
Unique NameID code of the higher taxon. Refers to the NameID code of LichenName (= foreign key). |
| TaxonNameCache |
nvarchar (500) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
| OrderNameID |
int |
NameID of the order |
| Order |
nvarchar (500) |
Name of the order |
Depending on:
Function HierarchySuperiorList
Returns a result set that lists all the taxa superior to a given taxon
| Parameter |
DataType |
Description |
| @NameID |
int |
Unique NameID code of the higher taxon. Refers to the NameID code of LichenName (= foreign key). |
| @ProjectID |
int |
Each project can implement a different taxonomic hierarchy. Refers to the common project definition in the DiversityProjects module. |
| Column |
DataType |
Description |
| NameID |
int |
Unique NameID code of the higher taxon. Refers to the NameID code of LichenName (= foreign key). |
| NameParentID |
int |
Next higher taxon (e.g. the family or subfamily if this taxon is a genus) |
| Seq |
int |
The sequence for the hierarchy starting with 1 for the Taxon corresponding to the given NameID |
| TaxonNameCache |
nchar (500) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
| Taxon |
nvarchar (500) |
Taxon excluding authors |
| TaxonomicRank |
nvarchar (50) |
Taxonomic rank of the taxon (var., subsp., species, genus, family, order, etc.). The rank must be selected from the associated list of ranks (= TaxonomyRank) |
| TaxonomicRankDisplayText |
nvarchar (50) |
The taxonomic rank in the case of a infraspecific taxon |
Depending on:
- TaxonHierarchy
- TaxonHierarchy_Core
- TaxonName
- TaxonNameTaxonomicRank_Enum
Function NameList
Returns a result set that lists all the taxa starting with a string corresponding to @NameStart and including the taxon corresponding to @NameID
| Parameter |
DataType |
Description |
| @NameID |
int |
The ID of a name in DiversityTaxonNames_Fungi (primary key) |
| @NameStart |
nvarchar (255) |
Initial string of the taxon (TaxonNameCache) |
| Column |
DataType |
Description |
| NameID |
int |
The ID of a name in DiversityTaxonNames_Fungi (primary key) |
| TaxonName |
nvarchar (200) |
Content of column TaxonNameCache |
Depending on:
Function NextNameID
Returns the next free NameID in the table TaxonName for the taxonomic group
DataType: int
| Parameter |
DataType |
Description |
| @InitialNameID |
int |
ID of the name where the query should start |
| @TaxonomicGroup |
nvarchar (50) |
TaxonomicGroup of the name. Ranges for the groups: |
| Biota: 0 |
|
|
| Myxomycetes: 1 - 99 999 |
|
|
| Plants: 100 000 - 999 999 |
|
|
| Lichens: 1 000 000 - 1 999 999 |
|
|
| Fungi: 2 000 000 - 2 999 999 |
|
|
| Vertebrata: 3 000 000 - 3 999 999 |
|
|
Depending on:
Function PrivacyConsentInfo
Website with information about the privacy consent. Default = http://diversityworkbench.net/Portal/Default_Agreement_on_Processing_of_Personal_Data_in_DWB_Software
DataType: varchar (900)
Function SynonymyList
Returns a result set that lists all the Synonyms for a given name
| Parameter |
DataType |
Description |
| @NameID |
int |
ID of the synonymized name. Refers to the NameID of TaxonName (= foreign key). |
| @ProjectID |
int |
ProjectID of the synonymy |
| @Level |
int |
the depth of the synonymy that should be requested. 0 = only linked via the basionym. Default = 3 |
| Column |
DataType |
Description |
| NameID |
int |
ID of the synonymized name. Refers to the NameID of TaxonName (= foreign key). |
| DisplayText |
nvarchar (900) |
A preformatet textstring for display in a synoymlist |
| TaxonName |
nvarchar (500) |
The name of the taxon including the genus and the species epithet |
| TaxonomicRank |
nvarchar (50) |
Taxonomic rank of the taxon (var., subsp., species, genus, family, order, etc.). The rank must be selected from the associated list of ranks (= TaxonomyRank) |
| InfraspecificEpithet |
nvarchar (200) |
The epithet of the infraspecific entity |
| Authors |
nvarchar (500) |
The combined authors of this name |
| SynonymText |
nvarchar (500) |
The text string containing the citation of the synonymisation |
| Prefix |
nvarchar (5) |
The prefix as used in synonym lists |
| Ord |
int |
The preferred sequence of the synonyms. The numbers in Ord correspond to the different kinds of the synonyms: |
| -20 = Accepted name |
|
|
| -10 = Basionym |
|
|
| 0 = Homotypic Synonym |
|
|
0 = Heterotypic Synoyms
x + 1 = Isonym resp. Assigned to like ‘duplicate’, ‘isonym’, ‘orthographic variant’ (will be display after the name it is a variant of) |
| YearOfPubl | int | The year when the name was published |
| TaxonNameCache | nvarchar (255) | A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
| ConceptSuffix | nvarchar (200) | Empty for default and primary concept; else ’s. lat.’, ’s. str.’, ‘sec.’, ‘sensu Muell.’, ’emend.’ , ‘(Auct.)’ etc. |
| ConceptNotes | nvarchar (500) | Notes on the concept of the name, e.g. incl. forms with black apothecia |
| BasedOnNameID | int | The Basionym of this name, resp. the NameID of the Basionym. A name is a Basionym, if NameID = BasionymID. |
| SynType | nvarchar (50) | Type of the synonymization, e.g. ‘heterotypic’, ‘homotypic’ |
| SynNameID | int | The ID of the synonym to which this name has been assigned to. Refers to the NameID of TaxonName (= foreign key). |
| NameParentID | int | The NameID of the parent within the synonymy |
Depending on:
- DefaultProjectID
- SynonymyListCache
- TaxonAcceptedName
- TaxonAcceptedNameList
- TaxonName
- TaxonSynonymy
- TaxonSynonymyList
Function SynonymyListIncludeIgnored
Returns a result set that lists all the Synonyms for a given name
| Parameter |
DataType |
Description |
| @NameID |
int |
ID of the accepted name. Refers to the NameID of TaxonName (= foreign key). |
| @ProjectID |
int |
Each project can have a different opinion regarding synonmy. Refers to the common project definition in the DiversityProjects module. |
| @Level |
int |
the depth of the synonymy that should be requested. 0 = only linked via the basionym. Default = 3 |
| Column |
DataType |
Description |
| NameID |
int |
ID of the accepted name. Refers to the NameID of TaxonName (= foreign key). |
| DisplayText |
nvarchar (900) |
A preformatet textstring for display in a synoymlist |
| TaxonName |
nvarchar (500) |
The name of the taxon including the genus and the species epithet |
| TaxonomicRank |
nvarchar (50) |
Taxonomic rank of the taxon (var., subsp., species, genus, family, order, etc.). The rank must be selected from the associated list of ranks (= TaxonomyRank) |
| InfraspecificEpithet |
nvarchar (200) |
The epithet of the infraspecific entity |
| Authors |
nvarchar (500) |
The combined authors of this name |
| SynonymText |
nvarchar (500) |
The text string containing the citation of the synonymisation |
| Prefix |
nvarchar (5) |
The prefix as used in synonym lists |
| Ord |
int |
The preferred sequence of the synonyms. The numbers in Ord correspond to the different kinds of the synonyms: |
| -20 = Accepted name |
|
|
| -10 = Basionym |
|
|
| 0 = Homotypic Synonym |
|
|
0 = Heterotypic Synoyms
x + 1 = Isonym resp. Assigned to like ‘duplicate’, ‘isonym’, ‘orthographic variant’ (will be display after the name it is a variant of) |
| YearOfPubl | int | The year when the name was published |
| TaxonNameCache | nvarchar (255) | A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
| ConceptSuffix | nvarchar (200) | Empty for default and primary concept; else ’s. lat.’, ’s. str.’, ‘sec.’, ‘sensu Muell.’, ’emend.’ , ‘(Auct.)’ etc. |
| ConceptNotes | nvarchar (500) | Notes on the concept of the name, e.g. incl. forms with black apothecia |
| BasedOnNameID | int | The Basionym of this name, resp. the NameID of the Basionym. A name is a Basionym, if NameID = BasionymID. |
| SynType | nvarchar (50) | Type of the synonymization, e.g. ‘heterotypic’, ‘homotypic’ |
| SynNameID | int | The ID of the synonym to which this name has been assigned to. Refers to the NameID of TaxonName (= foreign key). |
| NameParentID | int | - |
| IgnoreButKeepForReference | bit | If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead. |
Depending on:
- DefaultProjectID
- TaxonAcceptedName
- TaxonAcceptedNameListIncludeIgnored
- TaxonName
- TaxonSynonymy
- TaxonSynonymyListIncludeIgnored
Function TaxonChildNodes
Returns the list of taxa within the hierarchy underneath the taxon with the given NameID
| Parameter |
DataType |
Description |
| @NameID |
int |
The NameID of the taxon for which the hierarchy should be retrieved |
| @ProjectID |
int |
The ProjectID of the taxon for which the hierarchy should be retrieved |
| Column |
DataType |
Description |
| NameID |
int |
Unique NameID code of the higher taxon. Refers to the NameID code of LichenName (= foreign key). |
| TaxonNameCache |
nvarchar (255) |
A full unique version of the name. Generated by the database, not entered by the user (candidate key) |
| Version |
smallint |
The version of a name record (revision number, internally filled by system) |
| BasedOnNameID |
int |
The Basionym of this name, resp. the NameID of the Basionym. A name is a Basionym, if NameID = BasionymID. |
| CreationType |
nvarchar (50) |
E.g.: taxon based on new type, combination based on a previously publ. name (‘comb. nov.’), new name (’nom.nov.’) introduced to replace a homonym (may occur for genera!), validation of previously invalidly publ. taxon name (’ex’), or unknown. |
| TaxonomicRank |
nvarchar (50) |
Taxonomic rank of the taxon (var., subsp., species, genus, family, order, etc.). The rank must be selected from the associated list of ranks (= TaxonomyRank) |
| GenusOrSupragenericName |
nvarchar (200) |
If rank is above species: Name of taxon above species level (currently accepted spelling). Includes infrageneric taxon names, genera, families, etc. |
| SpeciesGenusNameID |
int |
If rank is species or below: NameID of the Genus name. Refers to same table with TaxonomicRank = genus. |
| InfragenericEpithet |
nvarchar (200) |
If rank below genus and above species: Name of infrageneric taxon above species level (currently accepted spelling). Includes subgenus, series, etc. |
| SpeciesEpithet |
nvarchar (100) |
The species name part of the species name, for example ‘alba’ in ‘Abies alba’. |
| InfraspecificEpithet |
nvarchar (100) |
The epithet of the infraspecific entity |
| BasionymAuthors |
nvarchar (100) |
The authors of a newly created name. For all taxonomic ranks, only for ‘comb. nov.’ or ’nom. nov.’: Author(s) of the basionym (will be displayed in ‘()’, do not enter the parentheses), abbreviated according to authors standard |
| CombiningAuthors |
nvarchar (255) |
The names of the combining authors if the name is base on another older name (e.g. combined into a different genus) |
| PublishingAuthors |
nvarchar (255) |
If the authors of the taxon differ from the authors of the publication: the latter (…‘in ’ Publishing authors … ) |
| SanctioningAuthor |
nvarchar (100) |
Sanctioning is a special instrument available for fungi to allow the starting date of taxonomy to be identical with higher plants, but preserve names used by Fries (Fr.) and Person (Pers.) |
| NonNomenclaturalNameSuffix |
nvarchar (200) |
A suffix for concept names not included in any nomenclatural code. If present fields for authors and typification must be empty and NomenclaturalCode set to ’not under code’. |
| IsRecombination |
bit |
True if the name is a recombination |
| IsHybrid |
bit |
If checked (or ‘True’), the new taxon is a hybrid with or without a hybrid epithet. |
| HybridNameID1 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 1. Refers to NomID code of publ. TaxonomyName (= foreign key). |
| HybridNameID2 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 2. Refers to NomID code of publ. TaxonomyName (= foreign key). |
| HybridNameID3 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 3. Refers to NomID code of publ. TaxonomyName (= foreign key). |
| HybridNameID4 |
int |
If IsHybrid is checked (or ‘True’): Name of hybrid species 4. Refers to NomID code of publ. TaxonomyName (= foreign key). |
| ReferenceTitle |
nvarchar (600) |
The title of the publication where the name was published. Note this is only a cached value where ReferenceURI is present |
| ReferenceURI |
varchar (255) |
URI (e.g. LSID) of Reference, referes to table ReferenceTitle in Database DiversityReferences: Source publication where name is published |
| Volume |
nvarchar (20) |
The volume of the journal |
| Issue |
nvarchar (255) |
The issue of the literature |
| Pages |
nvarchar (50) |
The pages within the literature |
| DetailLocation |
nvarchar (200) |
Additional information like plates etc. |
| DayOfPubl |
tinyint |
The day when the name was published |
| MonthOfPubl |
tinyint |
The month when the name was published |
| YearOfPubl |
smallint |
The year when the name was published |
| DateOfPublSupplement |
nvarchar (200) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
| YearOnPubl |
smallint |
The year cited on the original paper as year of publication |
| DateOnPublSupplement |
nvarchar (200) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
| Protologue |
nvarchar (MAX) |
Full text of the protologue. If protologue is provided in Latin and in another language |
| languages, both only the Latin or several languages may be stored together. |
|
|
| ProtologueURI |
varchar (255) |
URI of externally available Protologue information (scanned image, full text from external provider, etc.) |
| ProtologueResourceURI |
varchar (255) |
The ResourceID of an image of the protologue as stored in the module DiversityResources. Primary key of table Resource in the database DiversityResources.(= Foreign key) |
| NameUsageReferences |
nvarchar (255) |
Indexing volumes like Index of Fungi or any Name usage that caused this name to be added to the database |
|
|
|
| OriginalOrthography |
nvarchar (255) |
The original spelling of the name |
| NomenclaturalCode |
nvarchar (50) |
Code of Nomenclature under which this taxon was created: ‘Bacteriology’, ‘Botany’ (incl. Mycology), ‘Zoology’, ‘Biocode’ (for future use), ‘Non nomenclatural name’. Default value: ‘Botany’ |
| NomenclaturalStatus |
nvarchar (50) |
Categories for effective/valid/legitimate… esp. ’nom. illeg.’, ’nom. inval.’, ’nom. nudum’, etc. |
| NomenclaturalComment |
nvarchar (MAX) |
Comments on the nomenclature. e.g. ‘according to ICBN Art. 39.1’ |
| Typification |
nvarchar (50) |
The status of the type specimen(s) as written in the protologue, e.g. holotype |
| TypificationDetails |
nvarchar (255) |
Details concerning the typification as written in the protologue, e.g. if just parts of a specimen were accepted as the type |
| TypificationReferenceTitle |
nvarchar (255) |
The title of the publication where the typification was published. Note this is only a cached value where Typification-LiteratureLink is present |
| TypificationReferenceURI |
varchar (255) |
URI (e.g. LSID) of Reference, refers to e.g DiversityReferences: Source publication where the typification is published |
| TypificationNotes |
nvarchar (200) |
Notes concerning the typification |
| TypeSubstrate |
nvarchar (255) |
The substrate the type was growing on as written in the protologue |
| TypeLocality |
nvarchar (255) |
The locality where the type was found as written in the protologue |
| TypeSpecimenNotes |
nvarchar (200) |
Notes about the type specimen, e.g. the herbarium where a type specimen is located, Collector, collection date/number, etc. |
| AnamorphTeleomorph |
nvarchar (255) |
If the name is related to a anamorph or a teleomorph as written in the protologue |
| TypistNotes |
nvarchar (200) |
Additional notes and problems |
| RevisionLevel |
nvarchar (50) |
The level of the revision of the taxonomic name, e.g. ‘unchecked’, ‘fully checked’ |
| IgnoreButKeepForReference |
bit |
If true, the record is ignored for all purposes of evaluation (because contradicted). It is kept only to maintain the cited reference. If no reference is given, it may be deleted instead. |
| DataWithholdingReason |
nvarchar (255) |
If the dataset is withhold, the reason for withholding the data, otherwise null |
| BasionymAuthorsYear |
int |
The year when the name was published by the authors of the Basionym |
| FamiliyCache |
nvarchar (50) |
Name of the family |
| OrderCache |
nvarchar (50) |
Name of the order |
| HierarchyCache |
nvarchar (MAX) |
Hierarchy above the current taxon starting at top level and separated via " |
Depending on:
Function TaxonListLastUpdate
Returns the last date when any list related data were updated
DataType: datetime
| Parameter |
DataType |
Description |
| @ListID |
int |
ProjectID as defined in table TaxonNameListProjectProxy |
Depending on:
- TaxonNameList
- TaxonNameListAnalysis
- TaxonNameListArea
- TaxonNameListCollectionSpecimen
- TaxonNameListDistribution
- TaxonNameListReference
Function TaxonListsForUser
Returns the taxon lists for a user as defined in table TaxonNameListUser
| Parameter |
DataType |
Description |
| @Login |
nvarchar (50) |
Login of the user as stored in table TaxonNameListUser |
| Column |
DataType |
Description |
| ProjectID |
int |
ID of the Project, Primary key. May be related to module DiversityProjects |
| DataSource |
nvarchar (50) |
The project as defined in table TaxonNameListProjectProxy |
| DisplayText |
nvarchar (50) |
The text as shown in a user interface. Corresponds to ProjectTitle in DiversityProjects.Project |
| TaxonomicGroup |
nvarchar (50) |
The taxonomic group as used in DiversityCollection resp. DiversityMobile |
| DefaultProjectID |
int |
The ID of the default project where informations about e.g. the synonymy are taken from |
Depending on:
- TaxonNameListProjectProxy
- TaxonNameListUser
Function TaxonListsLastUpdate
Function providing informations needed by the webservice: Returns a result set that lists all the taxon lists and the date of their last change
| Column |
DataType |
Description |
| maxlog |
smalldatetime |
Date of the last change of a TaxonList |
| projectid |
int |
Each project can may contain one taxon list. Refers to the common project definition in the DiversityProjects module. |
Depending on:
- TaxonName
- TaxonName_Log
- TaxonNameList
Function TaxonNameCache
Returns the TaxonNameCache for a taxon
DataType: nvarchar (255)
| Parameter |
DataType |
Description |
| @NameID |
int |
The ID of a name in DiversityTaxonNames_Fungi (primary key) |
Depending on:
Function TaxonNameCacheSinAut
returns the TaxonName for a taxon excluding authors
DataType: nvarchar (255)
| Parameter |
DataType |
Description |
| @NameID |
int |
The ID of a name in DiversityTaxonNames_Fungi (primary key) |
Depending on:
Function TaxonNameListAnalysisCategoryChildNodes
Returns a result set that lists all analysis items within a hierarchy starting at the topmost item related to the given item
| Parameter |
DataType |
Description |
| @ID |
int |
AnalysisID of the analysis for which the child nodes should be returned |
| Column |
DataType |
Description |
| AnalysisID |
int |
ID of the analysis (Primary key) |
| AnalysisParentID |
int |
Analysis ID of the parent analysis if it belongs to a certain type documented in this table |
| DisplayText |
nvarchar (50) |
Name of the analysis as e.g. shown in user interface |
| Description |
nvarchar (500) |
Description of the analysis |
| Notes |
nvarchar (1000) |
Notes concerning this analysis |
| AnalysisURI |
varchar (255) |
URI referring to an external documentation of the analysis |
| ReferenceTitle |
nvarchar (800) |
The title of the publication where details about the analysis are published. Note this is only a cached value where ReferenceURI is present |
| ReferenceURI |
varchar (400) |
URI (e.g. LSID) of Reference, referes to table ReferenceTitle in module DiversityReferences: Source publication where details about the analysis are published |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
| SortingID |
int |
An ID for sorting the data |
Depending on:
- TaxonNameListAnalysisCategory
Function TaxonNameListAnalysisCategoryHierarchy
Returns a table that lists all the analysis items related to the given analysis
| Parameter |
DataType |
Description |
| @AnalysisID |
int |
ID of the analysis (Primary key) |
| Column |
DataType |
Description |
| AnalysisID |
int |
ID of the analysis (Primary key) |
| AnalysisParentID |
int |
Analysis ID of the parent analysis if it belongs to a certain type documented in this table |
| DisplayText |
nvarchar (50) |
Name of the analysis as e.g. shown in user interface |
| Description |
nvarchar (500) |
Description of the analysis |
| Notes |
nvarchar (1000) |
Notes concerning this analysis |
| AnalysisURI |
varchar (255) |
URI referring to an external documentation of the analysis |
| ReferenceTitle |
nvarchar (800) |
The title of the publication where details about the analysis are published. Note this is only a cached value where ReferenceURI is present |
| ReferenceURI |
varchar (400) |
URI (e.g. LSID) of Reference, referes to table ReferenceTitle in module DiversityReferences: Source publication where details about the analysis are published |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
| SortingID |
int |
An ID for sorting the data |
Depending on:
- TaxonNameListAnalysisCategory
- TaxonNameListAnalysisCategoryChildNodes
Function TaxonNameListUserNameID
Returns a result set that lists all the NameIDs for a user related to the taxon lists
| Column |
DataType |
Description |
| NameID |
int |
The ID of a name in DiversityTaxonNames_Fungi (primary key) |
Depending on:
- TaxonName
- TaxonNameList
- TaxonNameListUser
Function UserID
ID of the User as stored in table UserProxy
DataType: int
Depending on:
Function Version
Version of the database
DataType: nvarchar (8)
Function VersionClient
Version of the client compatible with the database
DataType: nvarchar (11)
PROCEDURES
Procedure procFillSynonymyListCache
Fills table SynonymyListCache for the given NameID or any if missing and ProjectID or any if missing
| Parameter |
DataType |
Description |
| @ProjectID |
int |
The ProjectID of the taxon for which the hierarchy should be retrieved |
| @NameID |
int |
The NameID of the taxon for which the hierarchy should be retrieved |
| @Level |
int |
The level how deep the hierarchy should extracted |
Depending on:
- ProjectProxy
- SynonymyList
- SynonymyListCache
- TaxonName
Procedure procHierarchyCache
Update of the hierarchy cache for all entries with disabled triggers to avoid increase of logfile entries
Depending on:
- HierarchyList
- TaxonHierarchy
Procedure procSetVersionTaxonName
Setting the value for column Version in table TaxonName
| Parameter |
DataType |
Description |
| @ID |
int |
The NameID of the taxon |
Depending on:
Procedure procTaxonNameInsertCopy
Copy all entries of a name into a new dataset
| Parameter |
DataType |
Description |
| @NameID |
int |
The ID of a name in DiversityTaxonNames (foreign key + part of primary key: the name may occur in multiple projects) |
| @OriginalNameID |
int |
The NameID of the dataset that should be copied |
| @Projects |
varchar (8000) |
List of comma separated ProjectIDs that should be copied |
Depending on:
- DefaultProjectID
- NextNameID
- ProjectList
- TaxonHierarchy_Core
- TaxonName_Core
- TaxonNameProject
Procedure procTaxonNameInsertNew_3
Inserts a new dataset in the table TaxonName
| Parameter |
DataType |
Description |
| @NameID |
int |
The ID of a name in DiversityTaxonNames_Fungi (primary key) |
| @Projects |
varchar (8000) |
Comma separated list of the project IDs for which the new taxon should be inserted |
| @NomenclaturalCode |
int |
Code of Nomenclature under which this taxon was created: ‘Bacteriology’, ‘Botany’ (incl. Mycology), ‘Zoology’, ‘Biocode’ (for future use), ‘Non nomenclatural name’. Default value: ‘Botany’ |
Depending on:
- TaxonName
- TaxonNameProject
Procedure procTaxonNameListInsertCopy
Copy all list entries of a name into the list of the current name
| Parameter |
DataType |
Description |
| @NameID |
int |
ID of the name. Refers to the NameID of TaxonName (= foreign key). |
| @OriginalNameID |
int |
The NameID of the dataset that should be copied |
| @ProjectID |
int |
Each project can may contain one taxon list. Refers to the common project definition in the DiversityProjects module. |
Depending on:
- TaxonNameList
- TaxonNameListAnalysis
- TaxonNameListCollectionSpecimen
- TaxonNameListDistribution
- TaxonNameListReference
Procedure procTaxonNameListInsertReferenceCopy
Copy all list entries of a name into the list of the current name
| Parameter |
DataType |
Description |
| @NameID |
int |
ID of the name. Refers to the NameID of TaxonName (= foreign key). |
| @O_TaxonNameListRefID |
int |
The PK of the reference that should be copied |
| @ProjectID |
int |
Each project can may contain one taxon list. Refers to the common project definition in the DiversityProjects module. |
Depending on:
- TaxonNameList
- TaxonNameListAnalysis
- TaxonNameListDistribution
- TaxonNameListReference
Procedure SetUserProjects
Create database user and assign training projects
| Parameter |
DataType |
Description |
| @User |
varchar (50) |
Login of the user |
Depending on:
ROLES
| Content of cell |
Permission |
|
Not granted |
| Name of other role |
Inherited from other role |
| • |
Granted |
Role DiversityWorkbenchAdministrator
Role for administration of the database content
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| CountryCode_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| LanguageCode_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| ProjectUser |
• |
• |
|
• |
|
TABLE |
| ReplicationPublisher |
• |
• |
• |
• |
|
TABLE |
| SynonymyListCache |
Diversity Workbench User |
Diversity Workbench Editor |
|
Diversity Workbench Editor |
|
TABLE |
| TaxonAcceptedName |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
TABLE |
| TaxonAcceptedName_log |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonCommonName |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
• |
|
TABLE |
| TaxonCommonName_log |
|
|
|
|
|
TABLE |
| TaxonGeography |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
TABLE |
| TaxonGeography_log |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonHierarchy |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
TABLE |
| TaxonHierarchy_log |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonName |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
• |
|
TABLE |
| TaxonName_log |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonNameCreationType_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameExternalDatabase |
Diversity Workbench User |
• |
• |
• |
|
TABLE |
| TaxonNameExternalDatabase_log |
|
|
|
|
|
TABLE |
| TaxonNameExternalID |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
• |
|
TABLE |
| TaxonNameExternalID_log |
|
|
|
|
|
TABLE |
| TaxonNameList |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
• |
|
TABLE |
| TaxonNameList_log |
Diversity Workbench User |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonNameListAnalysis |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
TABLE |
| TaxonNameListAnalysis_log |
Diversity Workbench User |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonNameListAnalysisCategory |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
• |
|
TABLE |
| TaxonNameListAnalysisCategory_log |
Diversity Workbench User |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonNameListAnalysisCategoryValue |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
• |
|
TABLE |
| TaxonNameListAnalysisCategoryValue_log |
Diversity Workbench User |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonNameListArea |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
TABLE |
| TaxonNameListArea_log |
Diversity Workbench User |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonNameListCollectionSpecimen |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
TABLE |
| TaxonNameListCollectionSpecimen_log |
|
|
|
|
|
TABLE |
| TaxonNameListDistribution |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
TABLE |
| TaxonNameListDistribution_log |
Diversity Workbench User |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonNameListImage |
|
|
|
|
|
TABLE |
| TaxonNameListImage_log |
|
|
|
|
|
TABLE |
| TaxonNameListReference |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
TABLE |
| TaxonNameListReference_log |
Diversity Workbench User |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonNameListReferenceType_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameListSpecimenRole_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameListUser |
Diversity Workbench User |
• |
|
• |
|
TABLE |
| TaxonNameNomenclaturalCode_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameNomenclaturalStatus_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameProject |
Diversity Workbench User |
Diversity Workbench Editor |
|
• |
|
TABLE |
| TaxonNameProject_log |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonNameReference |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
• |
|
TABLE |
| TaxonNameReference_log |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonNameResource |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
TABLE |
| TaxonNameResource_log |
Diversity Workbench User |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonNameResourceType_Enum |
Diversity Workbench User |
• |
• |
|
|
TABLE |
| TaxonNameRevisionLevel_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameSynonymisationType_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameTaxonomicRank_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameTypification |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
TABLE |
| TaxonNameTypification_log |
Diversity Workbench User |
Diversity Workbench Editor |
|
|
|
TABLE |
| TaxonSynonymy |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
TABLE |
| TaxonSynonymy_log |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
|
|
TABLE |
| AcceptedNameChanges |
Diversity Workbench User |
|
|
|
|
VIEW |
| GenusList |
|
|
|
|
|
VIEW |
| NameID_UserAvailable |
Diversity Workbench User |
|
|
|
|
VIEW |
| ProjectList |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonAcceptedName_Core |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
VIEW |
| TaxonAcceptedName_Indicated |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonAcceptedNameList |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonAcceptedNameListIncludeIgnored |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonFamily |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonGeography_Core |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
VIEW |
| TaxonHierarchy_Core |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
VIEW |
| TaxonListSynType |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonName_Core |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
• |
|
VIEW |
| TaxonName_Indicated |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
• |
|
VIEW |
| TaxonNameExternalID_Core |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonOrder |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonSynonymy_Core |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
VIEW |
| TaxonSynonymy_Indicated |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonSynonymyAcceptedNameID |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonSynonymyList |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonSynonymyListIncludeIgnored |
Diversity Workbench User |
|
|
|
|
VIEW |
| ViewBaseURL |
Diversity Workbench User |
|
|
|
|
VIEW |
| ViewDefaultProjectID |
Diversity Workbench User |
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
Diversity Workbench User |
|
|
|
|
VIEW |
| ViewTaxonNameResource |
Diversity Workbench User |
Diversity Workbench Editor |
Diversity Workbench Editor |
Diversity Workbench Editor |
|
VIEW |
| AcceptedNameID |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| AnalysisCategoryHierarchyAll |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| DefaultProjectID |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| DiversityMobile_TaxonList |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| HierarchyCache |
|
|
|
|
Diversity Workbench Editor |
FUNCTION |
| HierarchyFamilyList |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| HierarchyList |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| HierarchyOrderList |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| HierarchySuperiorList |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| NameList |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| NextNameID |
|
|
|
|
Diversity Workbench Editor |
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| SynonymyList |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| SynonymyListIncludeIgnored |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| TaxonChildNodes |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| TaxonListLastUpdate |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| TaxonListsForUser |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| TaxonListsLastUpdate |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| TaxonNameCache |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| TaxonNameCacheSinAut |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| TaxonNameListAnalysisCategoryChildNodes |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| TaxonNameListAnalysisCategoryHierarchy |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| TaxonNameListUserNameID |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| Version |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| VersionClient |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| procFillSynonymyListCache |
|
|
|
|
Diversity Workbench Editor |
PROCEDURE |
| procHierarchyCache |
|
|
|
|
Diversity Workbench Editor |
PROCEDURE |
| procSetVersionTaxonName |
|
|
|
|
|
PROCEDURE |
| procTaxonNameInsertCopy |
|
|
|
|
Diversity Workbench Editor |
PROCEDURE |
| procTaxonNameInsertNew_3 |
|
|
|
|
Diversity Workbench Editor |
PROCEDURE |
| procTaxonNameListInsertCopy |
|
|
|
|
Diversity Workbench Editor |
PROCEDURE |
| procTaxonNameListInsertReferenceCopy |
|
|
|
|
Diversity Workbench Editor |
PROCEDURE |
| SetUserProjects |
|
|
|
|
|
PROCEDURE |
| Inheriting from roles: |
|
|
|
|
|
|
Role DiversityWorkbenchEditor
Role for editing the user dependent content of the database
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| CountryCode_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| LanguageCode_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| ProjectUser |
Diversity Workbench User |
|
|
|
|
TABLE |
| ReplicationPublisher |
|
|
|
|
|
TABLE |
| SynonymyListCache |
Diversity Workbench User |
• |
|
• |
|
TABLE |
| TaxonAcceptedName |
Diversity Workbench User |
• |
• |
• |
|
TABLE |
| TaxonAcceptedName_log |
• |
• |
|
|
|
TABLE |
| TaxonCommonName |
Diversity Workbench User |
• |
• |
|
|
TABLE |
| TaxonCommonName_log |
|
|
|
|
|
TABLE |
| TaxonGeography |
Diversity Workbench User |
• |
• |
• |
|
TABLE |
| TaxonGeography_log |
• |
• |
|
|
|
TABLE |
| TaxonHierarchy |
Diversity Workbench User |
• |
• |
• |
|
TABLE |
| TaxonHierarchy_log |
• |
• |
|
|
|
TABLE |
| TaxonName |
Diversity Workbench User |
• |
• |
|
|
TABLE |
| TaxonName_log |
• |
• |
|
|
|
TABLE |
| TaxonNameCreationType_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameExternalDatabase |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameExternalDatabase_log |
|
|
|
|
|
TABLE |
| TaxonNameExternalID |
Diversity Workbench User |
• |
• |
• |
|
TABLE |
| TaxonNameExternalID_log |
|
|
|
|
|
TABLE |
| TaxonNameList |
Diversity Workbench User |
• |
• |
• |
|
TABLE |
| TaxonNameList_log |
Diversity Workbench User |
• |
|
|
|
TABLE |
| TaxonNameListAnalysis |
Diversity Workbench User |
• |
• |
• |
|
TABLE |
| TaxonNameListAnalysis_log |
Diversity Workbench User |
• |
|
|
|
TABLE |
| TaxonNameListAnalysisCategory |
Diversity Workbench User |
• |
• |
|
|
TABLE |
| TaxonNameListAnalysisCategory_log |
Diversity Workbench User |
• |
|
|
|
TABLE |
| TaxonNameListAnalysisCategoryValue |
Diversity Workbench User |
• |
• |
|
|
TABLE |
| TaxonNameListAnalysisCategoryValue_log |
Diversity Workbench User |
• |
|
|
|
TABLE |
| TaxonNameListArea |
Diversity Workbench User |
• |
• |
• |
|
TABLE |
| TaxonNameListArea_log |
Diversity Workbench User |
• |
|
|
|
TABLE |
| TaxonNameListCollectionSpecimen |
Diversity Workbench User |
• |
• |
• |
|
TABLE |
| TaxonNameListCollectionSpecimen_log |
|
|
|
|
|
TABLE |
| TaxonNameListDistribution |
Diversity Workbench User |
• |
• |
• |
|
TABLE |
| TaxonNameListDistribution_log |
Diversity Workbench User |
• |
|
|
|
TABLE |
| TaxonNameListImage |
|
|
|
|
|
TABLE |
| TaxonNameListImage_log |
|
|
|
|
|
TABLE |
| TaxonNameListReference |
Diversity Workbench User |
• |
• |
• |
|
TABLE |
| TaxonNameListReference_log |
Diversity Workbench User |
• |
|
|
|
TABLE |
| TaxonNameListReferenceType_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameListSpecimenRole_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameListUser |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameNomenclaturalCode_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameNomenclaturalStatus_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameProject |
Diversity Workbench User |
• |
|
|
|
TABLE |
| TaxonNameProject_log |
• |
• |
|
|
|
TABLE |
| TaxonNameReference |
Diversity Workbench User |
• |
• |
|
|
TABLE |
| TaxonNameReference_log |
• |
• |
|
|
|
TABLE |
| TaxonNameResource |
Diversity Workbench User |
• |
• |
• |
|
TABLE |
| TaxonNameResource_log |
Diversity Workbench User |
• |
|
|
|
TABLE |
| TaxonNameResourceType_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameRevisionLevel_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameSynonymisationType_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameTaxonomicRank_Enum |
Diversity Workbench User |
|
|
|
|
TABLE |
| TaxonNameTypification |
• |
• |
• |
• |
|
TABLE |
| TaxonNameTypification_log |
Diversity Workbench User |
• |
|
|
|
TABLE |
| TaxonSynonymy |
Diversity Workbench User |
• |
• |
• |
|
TABLE |
| TaxonSynonymy_log |
• |
• |
|
|
|
TABLE |
| AcceptedNameChanges |
Diversity Workbench User |
|
|
|
|
VIEW |
| GenusList |
|
|
|
|
|
VIEW |
| NameID_UserAvailable |
Diversity Workbench User |
|
|
|
|
VIEW |
| ProjectList |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonAcceptedName_Core |
Diversity Workbench User |
• |
• |
• |
|
VIEW |
| TaxonAcceptedName_Indicated |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonAcceptedNameList |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonAcceptedNameListIncludeIgnored |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonFamily |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonGeography_Core |
Diversity Workbench User |
• |
• |
• |
|
VIEW |
| TaxonHierarchy_Core |
Diversity Workbench User |
• |
• |
• |
|
VIEW |
| TaxonListSynType |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonName_Core |
Diversity Workbench User |
• |
• |
|
|
VIEW |
| TaxonName_Indicated |
Diversity Workbench User |
• |
• |
|
|
VIEW |
| TaxonNameExternalID_Core |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonOrder |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonSynonymy_Core |
Diversity Workbench User |
• |
• |
• |
|
VIEW |
| TaxonSynonymy_Indicated |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonSynonymyAcceptedNameID |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonSynonymyList |
Diversity Workbench User |
|
|
|
|
VIEW |
| TaxonSynonymyListIncludeIgnored |
Diversity Workbench User |
|
|
|
|
VIEW |
| ViewBaseURL |
Diversity Workbench User |
|
|
|
|
VIEW |
| ViewDefaultProjectID |
Diversity Workbench User |
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
Diversity Workbench User |
|
|
|
|
VIEW |
| ViewTaxonNameResource |
Diversity Workbench User |
• |
• |
• |
|
VIEW |
| AcceptedNameID |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| AnalysisCategoryHierarchyAll |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| DefaultProjectID |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| DiversityMobile_TaxonList |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| HierarchyCache |
|
|
|
|
• |
FUNCTION |
| HierarchyFamilyList |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| HierarchyList |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| HierarchyOrderList |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| HierarchySuperiorList |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| NameList |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| NextNameID |
|
|
|
|
• |
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| SynonymyList |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| SynonymyListIncludeIgnored |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| TaxonChildNodes |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| TaxonListLastUpdate |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| TaxonListsForUser |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| TaxonListsLastUpdate |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| TaxonNameCache |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| TaxonNameCacheSinAut |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| TaxonNameListAnalysisCategoryChildNodes |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| TaxonNameListAnalysisCategoryHierarchy |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| TaxonNameListUserNameID |
Diversity Workbench User |
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| Version |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| VersionClient |
|
|
|
|
Diversity Workbench User |
FUNCTION |
| procFillSynonymyListCache |
|
|
|
|
• |
PROCEDURE |
| procHierarchyCache |
|
|
|
|
• |
PROCEDURE |
| procSetVersionTaxonName |
|
|
|
|
|
PROCEDURE |
| procTaxonNameInsertCopy |
|
|
|
|
• |
PROCEDURE |
| procTaxonNameInsertNew_3 |
|
|
|
|
• |
PROCEDURE |
| procTaxonNameListInsertCopy |
|
|
|
|
• |
PROCEDURE |
| procTaxonNameListInsertReferenceCopy |
|
|
|
|
• |
PROCEDURE |
| SetUserProjects |
|
|
|
|
|
PROCEDURE |
| Inheriting from roles: |
|
|
|
|
|
|
Role DiversityWorkbenchReplicator
Role involved in the replication of data
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| CountryCode_Enum |
|
|
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| ProjectUser |
|
|
|
|
|
TABLE |
| ReplicationPublisher |
|
|
|
|
|
TABLE |
| SynonymyListCache |
|
|
|
|
|
TABLE |
| TaxonAcceptedName |
|
|
|
|
|
TABLE |
| TaxonAcceptedName_log |
|
|
|
|
|
TABLE |
| TaxonCommonName |
|
|
|
|
|
TABLE |
| TaxonCommonName_log |
|
|
|
|
|
TABLE |
| TaxonGeography |
|
|
|
|
|
TABLE |
| TaxonGeography_log |
|
|
|
|
|
TABLE |
| TaxonHierarchy |
|
|
|
|
|
TABLE |
| TaxonHierarchy_log |
|
|
|
|
|
TABLE |
| TaxonName |
|
|
|
|
|
TABLE |
| TaxonName_log |
|
|
|
|
|
TABLE |
| TaxonNameCreationType_Enum |
|
|
|
|
|
TABLE |
| TaxonNameExternalDatabase |
|
|
|
|
|
TABLE |
| TaxonNameExternalDatabase_log |
|
|
|
|
|
TABLE |
| TaxonNameExternalID |
|
|
|
|
|
TABLE |
| TaxonNameExternalID_log |
|
|
|
|
|
TABLE |
| TaxonNameList |
|
|
|
|
|
TABLE |
| TaxonNameList_log |
|
|
|
|
|
TABLE |
| TaxonNameListAnalysis |
|
|
|
|
|
TABLE |
| TaxonNameListAnalysis_log |
|
|
|
|
|
TABLE |
| TaxonNameListAnalysisCategory |
|
|
|
|
|
TABLE |
| TaxonNameListAnalysisCategory_log |
|
|
|
|
|
TABLE |
| TaxonNameListAnalysisCategoryValue |
|
|
|
|
|
TABLE |
| TaxonNameListAnalysisCategoryValue_log |
|
|
|
|
|
TABLE |
| TaxonNameListArea |
|
|
|
|
|
TABLE |
| TaxonNameListArea_log |
|
|
|
|
|
TABLE |
| TaxonNameListCollectionSpecimen |
|
|
|
|
|
TABLE |
| TaxonNameListCollectionSpecimen_log |
|
|
|
|
|
TABLE |
| TaxonNameListDistribution |
|
|
|
|
|
TABLE |
| TaxonNameListDistribution_log |
|
|
|
|
|
TABLE |
| TaxonNameListImage |
|
|
|
|
|
TABLE |
| TaxonNameListImage_log |
|
|
|
|
|
TABLE |
| TaxonNameListReference |
|
|
|
|
|
TABLE |
| TaxonNameListReference_log |
|
|
|
|
|
TABLE |
| TaxonNameListReferenceType_Enum |
|
|
|
|
|
TABLE |
| TaxonNameListSpecimenRole_Enum |
|
|
|
|
|
TABLE |
| TaxonNameListUser |
|
|
|
|
|
TABLE |
| TaxonNameNomenclaturalCode_Enum |
|
|
|
|
|
TABLE |
| TaxonNameNomenclaturalStatus_Enum |
|
|
|
|
|
TABLE |
| TaxonNameProject |
|
|
• |
|
|
TABLE |
| TaxonNameProject_log |
|
|
|
|
|
TABLE |
| TaxonNameReference |
|
|
|
|
|
TABLE |
| TaxonNameReference_log |
|
|
|
|
|
TABLE |
| TaxonNameResource |
|
|
|
|
|
TABLE |
| TaxonNameResource_log |
|
|
|
|
|
TABLE |
| TaxonNameResourceType_Enum |
|
|
|
|
|
TABLE |
| TaxonNameRevisionLevel_Enum |
|
|
|
|
|
TABLE |
| TaxonNameSynonymisationType_Enum |
|
|
|
|
|
TABLE |
| TaxonNameTaxonomicRank_Enum |
|
|
|
|
|
TABLE |
| TaxonNameTypification |
|
|
|
|
|
TABLE |
| TaxonNameTypification_log |
|
|
|
|
|
TABLE |
| TaxonSynonymy |
|
|
|
|
|
TABLE |
| TaxonSynonymy_log |
|
|
|
|
|
TABLE |
| AcceptedNameChanges |
|
|
|
|
|
VIEW |
| GenusList |
|
|
|
|
|
VIEW |
| NameID_UserAvailable |
|
|
|
|
|
VIEW |
| ProjectList |
|
|
|
|
|
VIEW |
| TaxonAcceptedName_Core |
|
|
|
|
|
VIEW |
| TaxonAcceptedName_Indicated |
|
|
|
|
|
VIEW |
| TaxonAcceptedNameList |
|
|
|
|
|
VIEW |
| TaxonAcceptedNameListIncludeIgnored |
|
|
|
|
|
VIEW |
| TaxonFamily |
|
|
|
|
|
VIEW |
| TaxonGeography_Core |
|
|
|
|
|
VIEW |
| TaxonHierarchy_Core |
|
|
|
|
|
VIEW |
| TaxonListSynType |
|
|
|
|
|
VIEW |
| TaxonName_Core |
|
|
|
|
|
VIEW |
| TaxonName_Indicated |
|
|
|
|
|
VIEW |
| TaxonNameExternalID_Core |
|
|
|
|
|
VIEW |
| TaxonOrder |
|
|
|
|
|
VIEW |
| TaxonSynonymy_Core |
|
|
|
|
|
VIEW |
| TaxonSynonymy_Indicated |
|
|
|
|
|
VIEW |
| TaxonSynonymyAcceptedNameID |
|
|
|
|
|
VIEW |
| TaxonSynonymyList |
|
|
|
|
|
VIEW |
| TaxonSynonymyListIncludeIgnored |
|
|
|
|
|
VIEW |
| ViewBaseURL |
|
|
|
|
|
VIEW |
| ViewDefaultProjectID |
|
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
|
|
|
|
|
VIEW |
| ViewTaxonNameResource |
|
|
|
|
|
VIEW |
| AcceptedNameID |
|
|
|
|
|
FUNCTION |
| AnalysisCategoryHierarchyAll |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
|
FUNCTION |
| DefaultProjectID |
|
|
|
|
|
FUNCTION |
| DiversityMobile_TaxonList |
|
|
|
|
|
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
|
FUNCTION |
| HierarchyCache |
|
|
|
|
|
FUNCTION |
| HierarchyFamilyList |
|
|
|
|
|
FUNCTION |
| HierarchyList |
|
|
|
|
|
FUNCTION |
| HierarchyOrderList |
|
|
|
|
|
FUNCTION |
| HierarchySuperiorList |
|
|
|
|
|
FUNCTION |
| NameList |
|
|
|
|
|
FUNCTION |
| NextNameID |
|
|
|
|
|
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
|
FUNCTION |
| SynonymyList |
|
|
|
|
|
FUNCTION |
| SynonymyListIncludeIgnored |
|
|
|
|
|
FUNCTION |
| TaxonChildNodes |
|
|
|
|
|
FUNCTION |
| TaxonListLastUpdate |
|
|
|
|
|
FUNCTION |
| TaxonListsForUser |
|
|
|
|
|
FUNCTION |
| TaxonListsLastUpdate |
|
|
|
|
|
FUNCTION |
| TaxonNameCache |
|
|
|
|
|
FUNCTION |
| TaxonNameCacheSinAut |
|
|
|
|
|
FUNCTION |
| TaxonNameListAnalysisCategoryChildNodes |
|
|
|
|
|
FUNCTION |
| TaxonNameListAnalysisCategoryHierarchy |
|
|
|
|
|
FUNCTION |
| TaxonNameListUserNameID |
|
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
|
FUNCTION |
| Version |
|
|
|
|
|
FUNCTION |
| VersionClient |
|
|
|
|
|
FUNCTION |
| procFillSynonymyListCache |
|
|
|
|
|
PROCEDURE |
| procHierarchyCache |
|
|
|
|
|
PROCEDURE |
| procSetVersionTaxonName |
|
|
|
|
|
PROCEDURE |
| procTaxonNameInsertCopy |
|
|
|
|
|
PROCEDURE |
| procTaxonNameInsertNew_3 |
|
|
|
|
|
PROCEDURE |
| procTaxonNameListInsertCopy |
|
|
|
|
|
PROCEDURE |
| procTaxonNameListInsertReferenceCopy |
|
|
|
|
|
PROCEDURE |
| SetUserProjects |
|
|
|
|
|
PROCEDURE |
Role DiversityWorkbenchUser
Role with read only access to the data
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| CountryCode_Enum |
• |
|
|
|
|
TABLE |
| LanguageCode_Enum |
• |
|
|
|
|
TABLE |
| ProjectUser |
• |
|
|
|
|
TABLE |
| ReplicationPublisher |
|
|
|
|
|
TABLE |
| SynonymyListCache |
• |
|
|
|
|
TABLE |
| TaxonAcceptedName |
• |
|
|
|
|
TABLE |
| TaxonAcceptedName_log |
|
|
|
|
|
TABLE |
| TaxonCommonName |
• |
|
|
|
|
TABLE |
| TaxonCommonName_log |
|
|
|
|
|
TABLE |
| TaxonGeography |
• |
|
|
|
|
TABLE |
| TaxonGeography_log |
|
|
|
|
|
TABLE |
| TaxonHierarchy |
• |
|
|
|
|
TABLE |
| TaxonHierarchy_log |
|
|
|
|
|
TABLE |
| TaxonName |
• |
|
|
|
|
TABLE |
| TaxonName_log |
|
|
|
|
|
TABLE |
| TaxonNameCreationType_Enum |
• |
|
|
|
|
TABLE |
| TaxonNameExternalDatabase |
• |
|
|
|
|
TABLE |
| TaxonNameExternalDatabase_log |
|
|
|
|
|
TABLE |
| TaxonNameExternalID |
• |
|
|
|
|
TABLE |
| TaxonNameExternalID_log |
|
|
|
|
|
TABLE |
| TaxonNameList |
• |
|
|
|
|
TABLE |
| TaxonNameList_log |
• |
|
|
|
|
TABLE |
| TaxonNameListAnalysis |
• |
|
|
|
|
TABLE |
| TaxonNameListAnalysis_log |
• |
|
|
|
|
TABLE |
| TaxonNameListAnalysisCategory |
• |
|
|
|
|
TABLE |
| TaxonNameListAnalysisCategory_log |
• |
|
|
|
|
TABLE |
| TaxonNameListAnalysisCategoryValue |
• |
|
|
|
|
TABLE |
| TaxonNameListAnalysisCategoryValue_log |
• |
|
|
|
|
TABLE |
| TaxonNameListArea |
• |
|
|
|
|
TABLE |
| TaxonNameListArea_log |
• |
|
|
|
|
TABLE |
| TaxonNameListCollectionSpecimen |
• |
|
|
|
|
TABLE |
| TaxonNameListCollectionSpecimen_log |
|
|
|
|
|
TABLE |
| TaxonNameListDistribution |
• |
|
|
|
|
TABLE |
| TaxonNameListDistribution_log |
• |
|
|
|
|
TABLE |
| TaxonNameListImage |
|
|
|
|
|
TABLE |
| TaxonNameListImage_log |
|
|
|
|
|
TABLE |
| TaxonNameListReference |
• |
|
|
|
|
TABLE |
| TaxonNameListReference_log |
• |
|
|
|
|
TABLE |
| TaxonNameListReferenceType_Enum |
• |
|
|
|
|
TABLE |
| TaxonNameListSpecimenRole_Enum |
• |
|
|
|
|
TABLE |
| TaxonNameListUser |
• |
|
|
|
|
TABLE |
| TaxonNameNomenclaturalCode_Enum |
• |
|
|
|
|
TABLE |
| TaxonNameNomenclaturalStatus_Enum |
• |
|
|
|
|
TABLE |
| TaxonNameProject |
• |
|
|
|
|
TABLE |
| TaxonNameProject_log |
|
|
|
|
|
TABLE |
| TaxonNameReference |
• |
|
|
|
|
TABLE |
| TaxonNameReference_log |
|
|
|
|
|
TABLE |
| TaxonNameResource |
• |
|
|
|
|
TABLE |
| TaxonNameResource_log |
• |
|
|
|
|
TABLE |
| TaxonNameResourceType_Enum |
• |
|
|
|
|
TABLE |
| TaxonNameRevisionLevel_Enum |
• |
|
|
|
|
TABLE |
| TaxonNameSynonymisationType_Enum |
• |
|
|
|
|
TABLE |
| TaxonNameTaxonomicRank_Enum |
• |
|
|
|
|
TABLE |
| TaxonNameTypification |
• |
|
|
|
|
TABLE |
| TaxonNameTypification_log |
• |
|
|
|
|
TABLE |
| TaxonSynonymy |
• |
|
|
|
|
TABLE |
| TaxonSynonymy_log |
|
|
|
|
|
TABLE |
| AcceptedNameChanges |
• |
|
|
|
|
VIEW |
| GenusList |
|
|
|
|
|
VIEW |
| NameID_UserAvailable |
• |
|
|
|
|
VIEW |
| ProjectList |
• |
|
|
|
|
VIEW |
| TaxonAcceptedName_Core |
• |
|
|
|
|
VIEW |
| TaxonAcceptedName_Indicated |
• |
|
|
|
|
VIEW |
| TaxonAcceptedNameList |
• |
|
|
|
|
VIEW |
| TaxonAcceptedNameListIncludeIgnored |
• |
|
|
|
|
VIEW |
| TaxonFamily |
• |
|
|
|
|
VIEW |
| TaxonGeography_Core |
• |
|
|
|
|
VIEW |
| TaxonHierarchy_Core |
• |
|
|
|
|
VIEW |
| TaxonListSynType |
• |
|
|
|
|
VIEW |
| TaxonName_Core |
• |
|
|
|
|
VIEW |
| TaxonName_Indicated |
• |
|
|
|
|
VIEW |
| TaxonNameExternalID_Core |
• |
|
|
|
|
VIEW |
| TaxonOrder |
• |
|
|
|
|
VIEW |
| TaxonSynonymy_Core |
• |
|
|
|
|
VIEW |
| TaxonSynonymy_Indicated |
• |
|
|
|
|
VIEW |
| TaxonSynonymyAcceptedNameID |
• |
|
|
|
|
VIEW |
| TaxonSynonymyList |
• |
|
|
|
|
VIEW |
| TaxonSynonymyListIncludeIgnored |
• |
|
|
|
|
VIEW |
| ViewBaseURL |
• |
|
|
|
|
VIEW |
| ViewDefaultProjectID |
• |
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
• |
|
|
|
|
VIEW |
| ViewTaxonNameResource |
• |
|
|
|
|
VIEW |
| AcceptedNameID |
|
|
|
|
• |
FUNCTION |
| AnalysisCategoryHierarchyAll |
• |
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
• |
FUNCTION |
| DefaultProjectID |
|
|
|
|
• |
FUNCTION |
| DiversityMobile_TaxonList |
• |
|
|
|
|
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
• |
FUNCTION |
| HierarchyCache |
|
|
|
|
|
FUNCTION |
| HierarchyFamilyList |
• |
|
|
|
|
FUNCTION |
| HierarchyList |
• |
|
|
|
|
FUNCTION |
| HierarchyOrderList |
• |
|
|
|
|
FUNCTION |
| HierarchySuperiorList |
• |
|
|
|
|
FUNCTION |
| NameList |
• |
|
|
|
|
FUNCTION |
| NextNameID |
|
|
|
|
|
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
• |
FUNCTION |
| SynonymyList |
• |
|
|
|
|
FUNCTION |
| SynonymyListIncludeIgnored |
• |
|
|
|
|
FUNCTION |
| TaxonChildNodes |
• |
|
|
|
|
FUNCTION |
| TaxonListLastUpdate |
|
|
|
|
• |
FUNCTION |
| TaxonListsForUser |
• |
|
|
|
|
FUNCTION |
| TaxonListsLastUpdate |
• |
|
|
|
|
FUNCTION |
| TaxonNameCache |
|
|
|
|
• |
FUNCTION |
| TaxonNameCacheSinAut |
|
|
|
|
• |
FUNCTION |
| TaxonNameListAnalysisCategoryChildNodes |
• |
|
|
|
|
FUNCTION |
| TaxonNameListAnalysisCategoryHierarchy |
• |
|
|
|
|
FUNCTION |
| TaxonNameListUserNameID |
• |
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
• |
FUNCTION |
| Version |
|
|
|
|
• |
FUNCTION |
| VersionClient |
|
|
|
|
• |
FUNCTION |
| procFillSynonymyListCache |
|
|
|
|
|
PROCEDURE |
| procHierarchyCache |
|
|
|
|
|
PROCEDURE |
| procSetVersionTaxonName |
|
|
|
|
|
PROCEDURE |
| procTaxonNameInsertCopy |
|
|
|
|
|
PROCEDURE |
| procTaxonNameInsertNew_3 |
|
|
|
|
|
PROCEDURE |
| procTaxonNameListInsertCopy |
|
|
|
|
|
PROCEDURE |
| procTaxonNameListInsertReferenceCopy |
|
|
|
|
|
PROCEDURE |
| SetUserProjects |
|
|
|
|
|
PROCEDURE |
Diversity TaxonNames
No-SQL Interface
JSON Cache
Databases of the modules provid a No-SQL interface where the data of the main table and the depending tables are merged as JSON.
Table JsonCache
Content of table JsonCache restricted to public available data
| Column |
Data type |
Description |
Nullable |
Relation |
| ID |
int |
Unique ID for the Dataset, Primary key |
NO |
ID of the source |
| URI |
varchar (500) |
The URL as combination of BaseURL and ID |
NO |
BaseURL and ID of the source |
| DisplayText |
nvarchar (500) |
Representation in the interface |
NO |
Main table of the source |
| LogUpdatedWhen |
datetime |
Date and time when the data were last updated |
NO |
- |
| Data |
json |
Data related to the current dataset |
NO |
- |
procFillJsonCache is started by an update trigger trgUpd… of the main table in the database
Interface in clients
All modules provide data via a cache table. In the header of the clients you can inspect the content of the JsonCache with a click on the  button. For modules with a difference between local and public data, you can inspect the content of the public data with a click of the right mouse button.
button. For modules with a difference between local and public data, you can inspect the content of the public data with a click of the right mouse button.
Update
Apart of the update via the Trigger (see below) you can update the JsonCache via the  update button underneath the
update button underneath the  button.
button.
To update the JsonCache for the whole database select Administration - JsonCache… from the menu. a window as shown below will open where you can update the JsonCache for single datasets or the whole database.
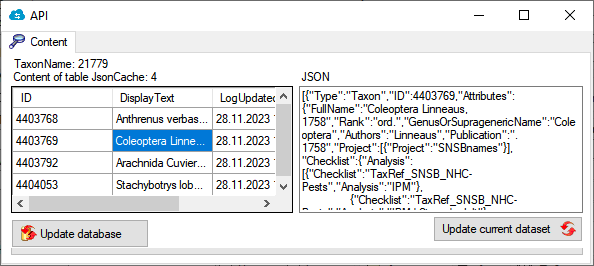
Summary
graph TD;
TaxonName[Main table in database]
trgUpdTaxonName[trgUpd.. of main table in database]
TaxonName --> |Update in table| trgUpdTaxonName
proc[Procedure procFillJsonCache setting the content in table JsonCache]
trgUpdTaxonName --> proc
graph TD;
Mainform[Main form]
ButtonShow[Button show JsonCache of current dataset]
Mainform --> ButtonShow
Left[Show Data]
ButtonShow --> |Left click| Left
graph TD;
Mainform[Main form]
Admin[Administration menu]
Mainform --> Admin
Cache[JsonCache...]
Admin --> Cache
Adminform[Administration form]
Cache --> Adminform
AdminUpdateSingle[Update single dataset]
Adminform --> AdminUpdateSingle
AdminUpdateDB[Update for whole database]
Adminform --> AdminUpdateDB
Example
The JsonCache of the current dataset will be shown as in the example below:
[
{
"Type": "Taxon",
"ID": 4403792,
"Attributes": {
"FullName": "Arachnida Cuvier, 1812",
"Rank": "cl.",
"GenusOrSupragenericName": "Arachnida",
"Authors": "Cuvier",
"Publication": ". 1812",
"Project": [
{
"Project": "SNSBnames"
}
],
"Checklist": {
"Analysis": [
{
"Checklist": "TaxRef_SNSB_NHC-Pests",
"Analysis": "IPM | Group",
"Value": "70"
},
{
"Checklist": "TaxRef_SNSB_NHC-Beneficials",
"Analysis": "IPM | Group",
"Value": "70"
},
{
"Checklist": "TaxRef_SNSB_NHC-Bycatchs",
"Analysis": "IPM | Group",
"Value": "70"
}
]
},
"Synonymy": [
{
"ID": 4403792,
"FullName": "Arachnida Cuvier, 1812",
"Name": "Arachnida",
"Type": "accepted name"
}
],
"HierarchyListCache": [
{
"HierarchyListCache": "Animalia | Arthropoda | Arachnida"
}
],
"Hierarchy": [
{
"ID": 4403792,
"FullName": "Arachnida Cuvier, 1812",
"Name": "Arachnida",
"Rank": "cl."
},
{
"ID": 4403934,
"FullName": "Arthropoda Gravenhorst, 1843",
"Name": "Arthropoda",
"Rank": "phyl./div."
},
{
"ID": 4403805,
"FullName": "Animalia Linnaeus, 1758",
"Name": "Animalia",
"Rank": "reg."
}
],
"CommonNames": [
{
"CommonName": "spider",
"LanguageCode": "en",
"CountryCode": "GB",
"Context": "IPM"
},
{
"CommonName": "Spinnentier",
"LanguageCode": "de",
"CountryCode": "DE",
"Context": "IPM"
}
],
"Resource": [
{
"URI": "https://museumsschaedlinge.de/gaeste/",
"Title": "diverse animals that migrate or are introduced into buildings seasonally, generally don't casue any damage to objects",
"Type": "information"
}
]
}
}
]
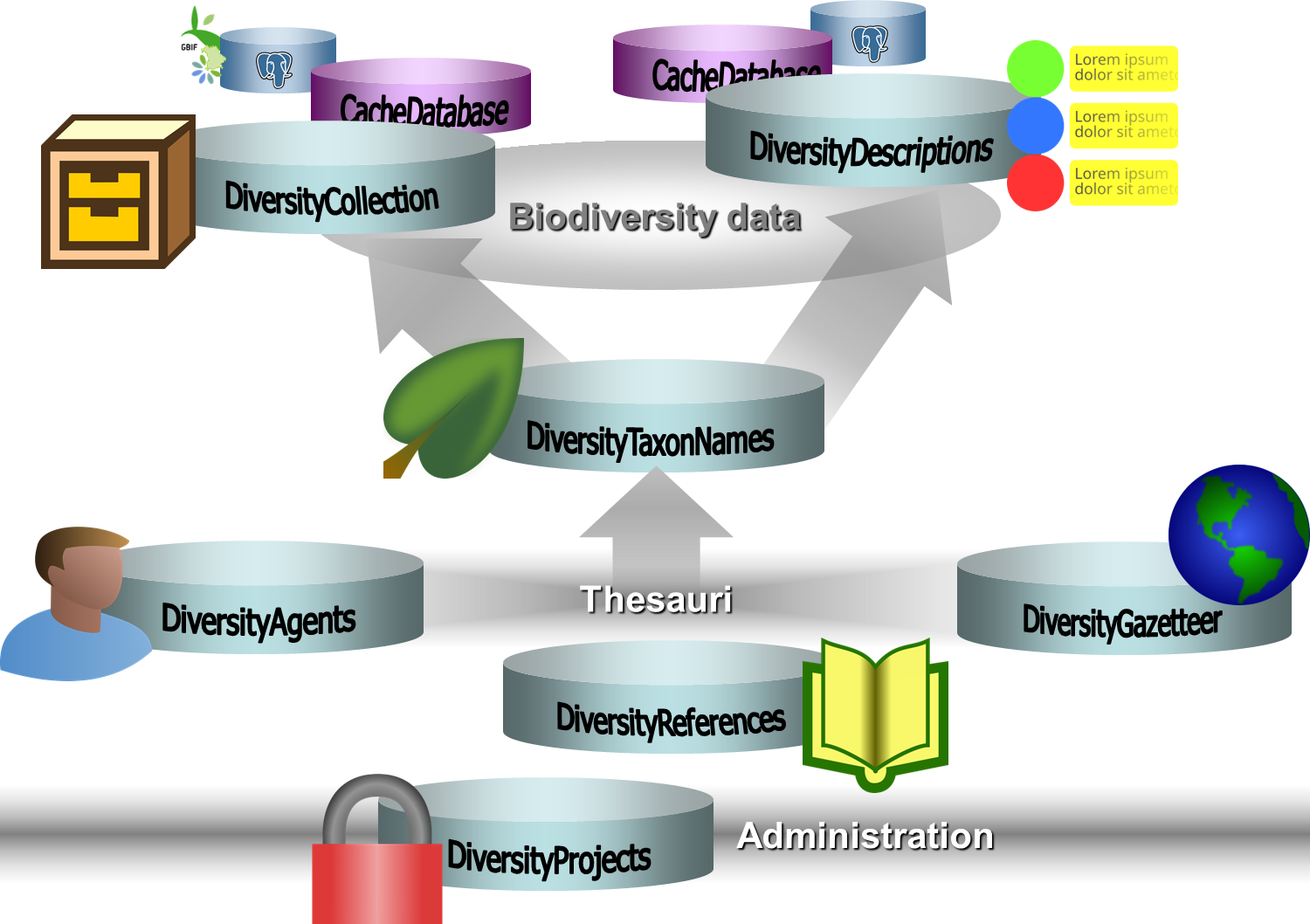 The modules communicate with each other to provide their
services for the other modules.
The modules communicate with each other to provide their
services for the other modules.

 New table JsonCache containing the data related to an agent as JSON. In the menu accessible via Administration →
New table JsonCache containing the data related to an agent as JSON. In the menu accessible via Administration →  Bugfix updating linked datasets, svn 6528,
Bugfix updating linked datasets, svn 6528,  Show Images for analysis value in case the value is a link to an image
Show Images for analysis value in case the value is a link to an image Hierarchy
Hierarchy
 Project
Project
 ProjectUser: Adding ReadOnly for locking a project
ProjectUser: Adding ReadOnly for locking a project New function ProjectDataLastChanges
New function ProjectDataLastChanges ProjectProxy: Adding missing columns for archive
ProjectProxy: Adding missing columns for archive Missing descriptions
Missing descriptions TaxonNameListAnalysisCategory: Adding parent relation
TaxonNameListAnalysisCategory: Adding parent relation Refreshing the hierarchy: Warning when changing the root of the displayed hierarchy
Refreshing the hierarchy: Warning when changing the root of the displayed hierarchy



 DiversityCollection.exe.
DiversityCollection.exe.

 Database …: Choose one of the databases available on the server. Only those
databases will be listed to which the user has access permission
Database …: Choose one of the databases available on the server. Only those
databases will be listed to which the user has access permission Module connections …: Edit the connections to the other modules within the Diversity Workbench.
Module connections …: Edit the connections to the other modules within the Diversity Workbench. Transfer previous settings: Transfer the settings for IP-Address and port of the server, name of the database, login etc. of a previous version of the client to the current version.
Transfer previous settings: Transfer the settings for IP-Address and port of the server, name of the database, login etc. of a previous version of the client to the current version. Quit: Quit the application and stop all processes started by the application
Quit: Quit the application and stop all processes started by the application View taxa in table …: Show the taxa in a spreadsheet
View taxa in table …: Show the taxa in a spreadsheet Edit taxa in table …: Edit the taxa in a spreadsheet
Edit taxa in table …: Edit the taxa in a spreadsheet Spreadsheets: Edit data in configurable tables.
Spreadsheets: Edit data in configurable tables.
 Taxon sheet…: Edit taxon and dependent data.
Taxon sheet…: Edit taxon and dependent data. Common name…: Edit data in table TaxonCommonName.
Common name…: Edit data in table TaxonCommonName. Typification…: Edit data in table TaxonNameTypification.
Typification…: Edit data in table TaxonNameTypification. Geography…: Edit data in table TaxonGeography.
Geography…: Edit data in table TaxonGeography. Accepted name…: Edit data in table TaxonAcceptedName.
Accepted name…: Edit data in table TaxonAcceptedName. Synonymy…: Edit data in table TaxonSynonymy.
Synonymy…: Edit data in table TaxonSynonymy. Hierarchy…: Edit data in table TaxonHierarchy.
Hierarchy…: Edit data in table TaxonHierarchy. List: Edit data in tables depending on list.
List: Edit data in tables depending on list.
 Specimen…: Edit data in table TaxonNameListCollectionSpecimen.
Specimen…: Edit data in table TaxonNameListCollectionSpecimen. Reference…: Edit data in table TaxonNameListReference.
Reference…: Edit data in table TaxonNameListReference. Distribution…: Edit data in table TaxonNameListDistribution.
Distribution…: Edit data in table TaxonNameListDistribution. Import
Import
 Import wizard: Import data from tab-separated text files
Import wizard: Import data from tab-separated text files
 Webservices: Import taxonomic names from webservices
Webservices: Import taxonomic names from webservices MYCOBANK…: Import taxa from MYCOBANK
MYCOBANK…: Import taxa from MYCOBANK Export
Export
 Export wizard …: Export tab-separated text files
Export wizard …: Export tab-separated text files
 Export as CSV (bcp): Export data of the whole database as csv files
Export as CSV (bcp): Export data of the whole database as csv files Replication …: Synchronise the content of 2 databases.
Replication …: Synchronise the content of 2 databases.
 Add publisher …: Add a replication publisher for the replication.
Add publisher …: Add a replication publisher for the replication. Download …: Download data from the data provider in your local database.
Download …: Download data from the data provider in your local database. Merge …: Merge contents between your local database and the replication provider.
Merge …: Merge contents between your local database and the replication provider. Upload …: Upload data from your local database to the replication provider.
Upload …: Upload data from your local database to the replication provider. Remove: Remove the replication provider.
Remove: Remove the replication provider. Clean database …: Clean your local database.
Clean database …: Clean your local database. Create archive…: Create and archive of project data.
Create archive…: Create and archive of project data. Restore archive…: Restore project data from an archive.
Restore archive…: Restore project data from an archive. Backup database …: Backup of the whole database
Backup database …: Backup of the whole database Remove taxa…: Remove all taxa selected in the query
Remove taxa…: Remove all taxa selected in the query Logins …: Administration of the logins and user
Logins …: Administration of the logins and user Maintenance…: Maintenace functions within the DiversityTaxonNames
Maintenance…: Maintenace functions within the DiversityTaxonNames Rename database: Rename the current database
Rename database: Rename the current database Database tools: Tools to create and change objects in the database
Database tools: Tools to create and change objects in the database Manual: Opens the online manual
Manual: Opens the online manual Feedback…: Opens a window for sending feedback
Feedback…: Opens a window for sending feedback Feedback history…: Opens a window for browsing former feedback
Feedback history…: Opens a window for browsing former feedback Statistics: Show the statistics within a project
Statistics: Show the statistics within a project Info: Show the version and corresponding information
Info: Show the version and corresponding information Chart: Opens a chart query
Chart: Opens a chart query
 .
.
 .
.
 .
. .
. .
. , Problem
, Problem  , Reference
, Reference
 . Remove a ‘duplicate’ search condition with a click on the red “Minus” button
. Remove a ‘duplicate’ search condition with a click on the red “Minus” button 
 .
.

 . To display
several columns in the result list, click on the
. To display
several columns in the result list, click on the  button.
To change the sorting of the added order column click on the
button.
To change the sorting of the added order column click on the

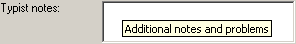
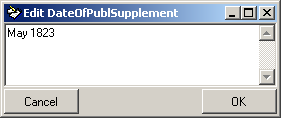
 and
and  button to open the corresponding website in a
separate window.
button to open the corresponding website in a
separate window.


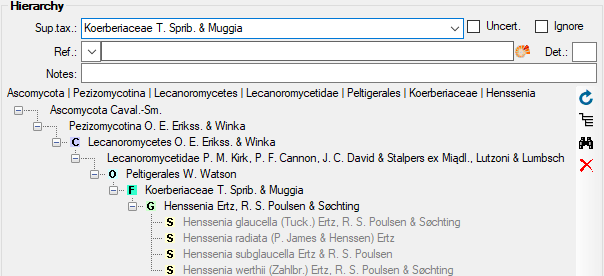
 . The button
. The button
 button.
button.

 button for an optimal height of the rows or
the
button for an optimal height of the rows or
the  button for an optimal width of the columns.
button for an optimal width of the columns. (= Append). The
(= Append). The  botton opens
the folder containing the exported file.
botton opens
the folder containing the exported file.

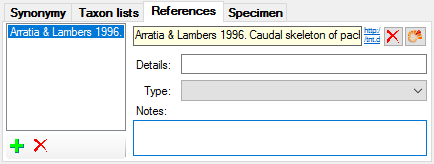

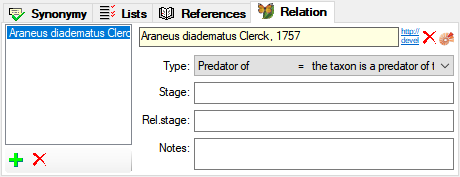


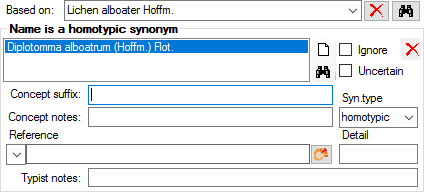
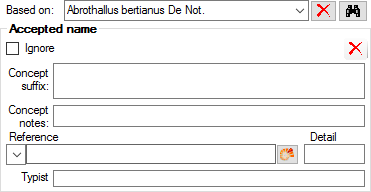 You can indicate
accepted names in the search result list (in the menu choose View,
indicate accepted names)
You can indicate
accepted names in the search result list (in the menu choose View,
indicate accepted names)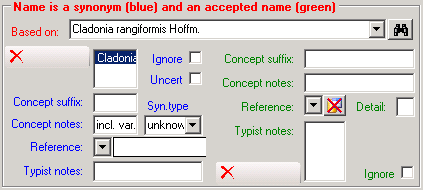
 button to change between these views. In the tree view the current name
is marked with a yellow background as shown below. The scrutiny of the search can be
adapted by changing the number in the tool bar
button to change between these views. In the tree view the current name
is marked with a yellow background as shown below. The scrutiny of the search can be
adapted by changing the number in the tool bar  save button next to the basionym to update the
synonymy in the cache table for the current name or choose Administation -
save button next to the basionym to update the
synonymy in the cache table for the current name or choose Administation - 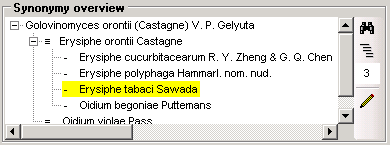

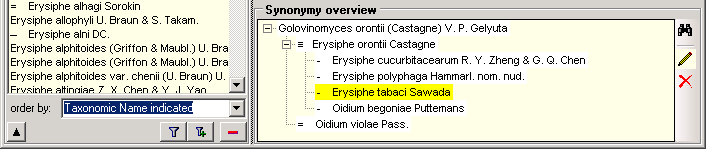

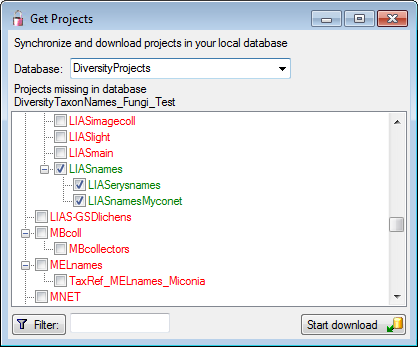
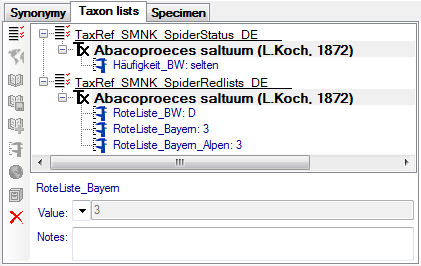
 button and search for the region as described in
the chapter
button and search for the region as described in
the chapter 


 button. As a result you
can use the
button. As a result you
can use the  button to insert these
informations at a different place.
button to insert these
informations at a different place. . The sorting in an interface is set via the
sorting. To check the current sorting click on the
. The sorting in an interface is set via the
sorting. To check the current sorting click on the 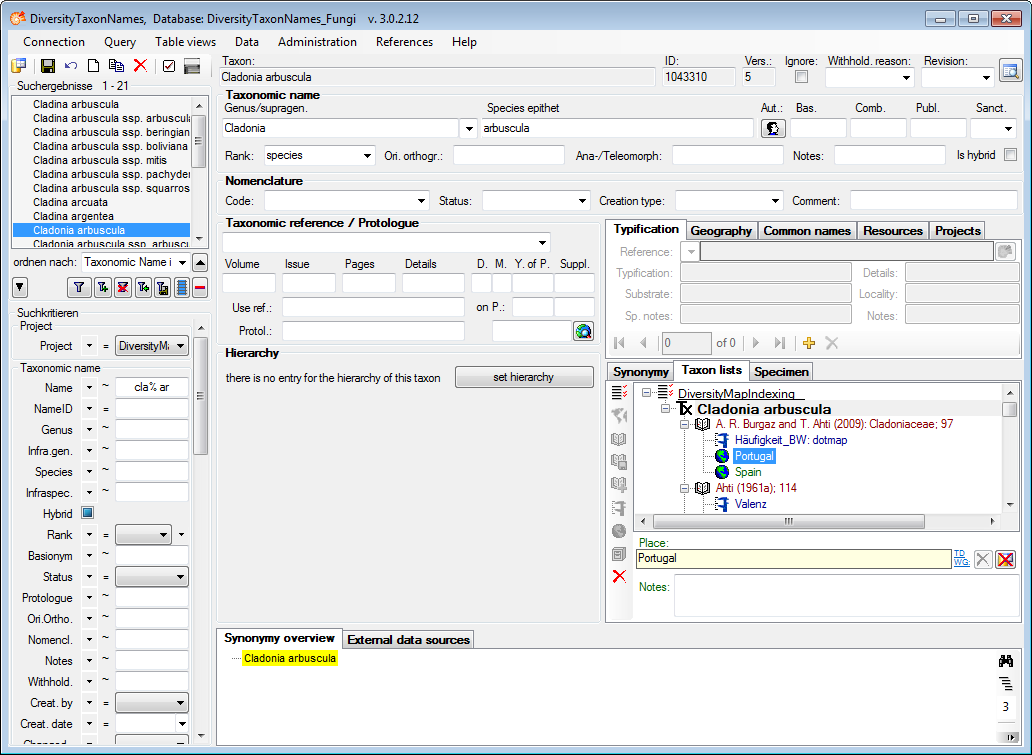
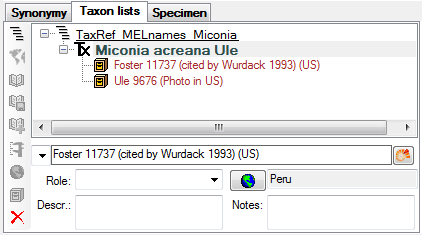
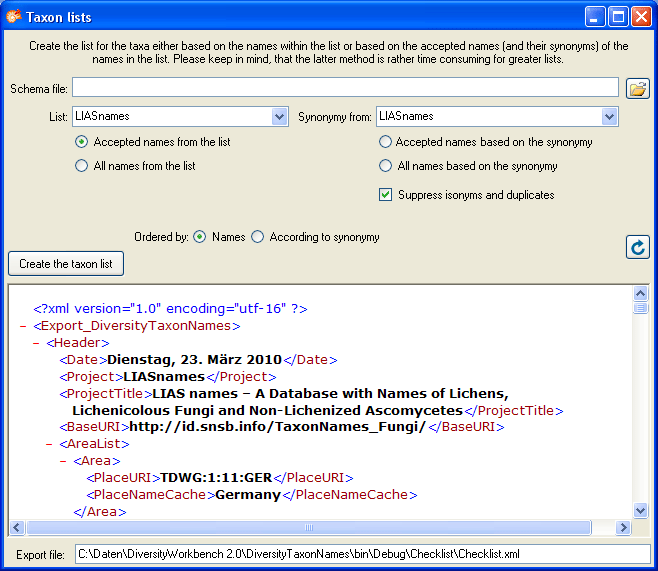
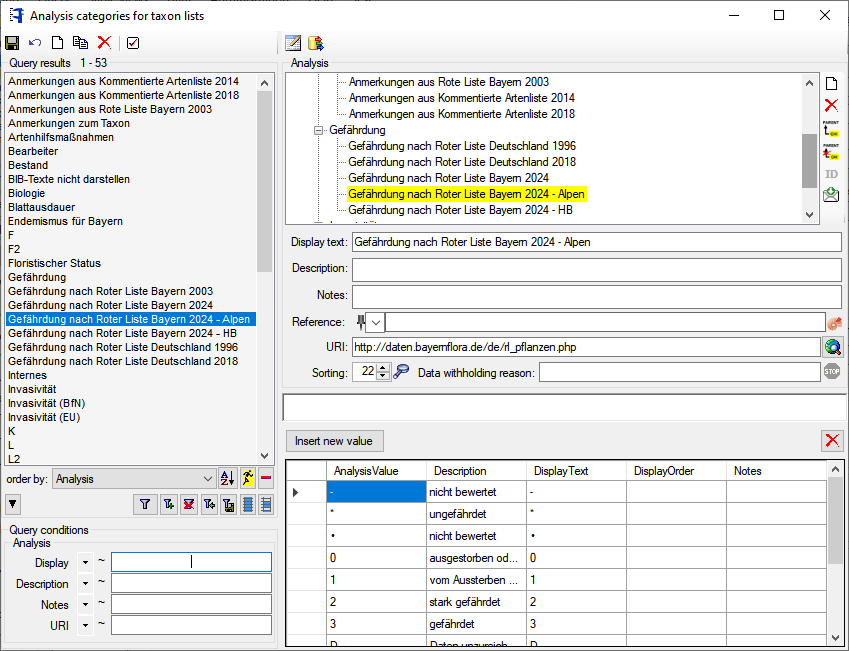

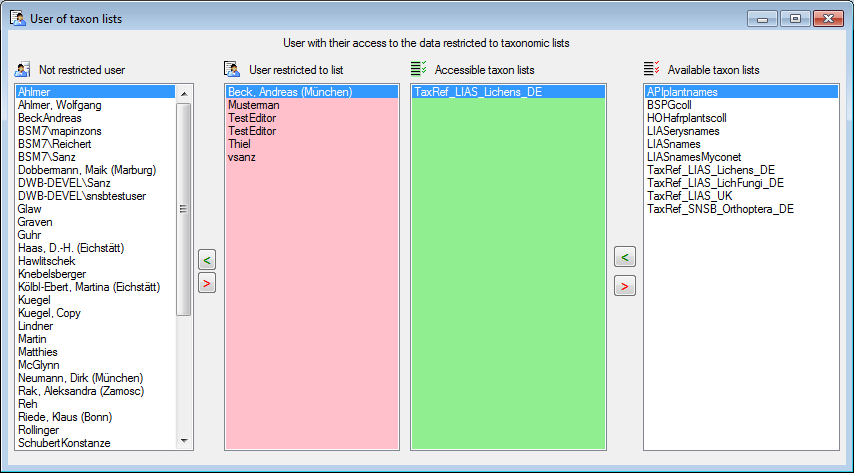
 undo button. This will
recover the original data unless the changes had been saved or changes
were done in the synonymy.
undo button. This will
recover the original data unless the changes had been saved or changes
were done in the synonymy.
 . As with the
creation of a new dataset you can insert the copy of a name into other
projects (see above).
. As with the
creation of a new dataset you can insert the copy of a name into other
projects (see above). button.
button.
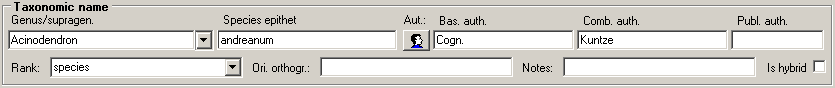
 and the
program will not allow you to edit the name. To edit the genus of the
name, click the
and the
program will not allow you to edit the name. To edit the genus of the
name, click the 
 Button. This will open the form for editing the authors.
Button. This will open the form for editing the authors.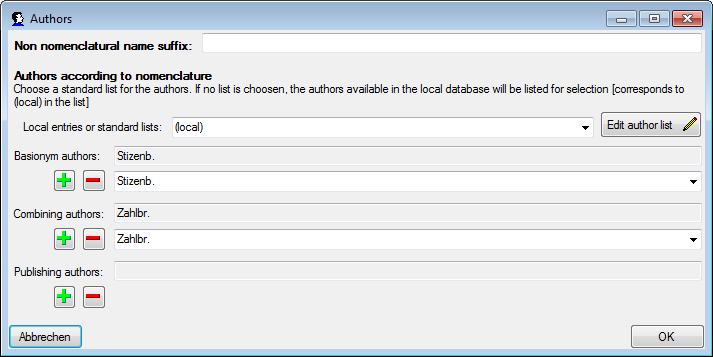

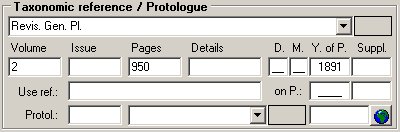
 ,
,
 ,
,  and
and
 button. To add a new typification use the
button. To add a new typification use the

 Ignore option. All data set to
Ignore option. All data set to
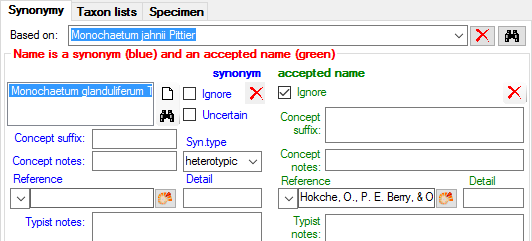

 . From
there they are transferred into a
. From
there they are transferred into a  containing a package for conversion into
ABCD. Finally the
containing a package for conversion into
ABCD. Finally the BioCASE tool for mapping the
data is used to provide the data for GBIF.
BioCASE tool for mapping the
data is used to provide the data for GBIF.
 option as described in the tutorial:
option as described in the tutorial: 
 Restore archive... from the menu. A
window as shown below will open listing the tables in the database. To
restore an archive click on the Choose archive directory
Restore archive... from the menu. A
window as shown below will open listing the tables in the database. To
restore an archive click on the Choose archive directory
 option as described in the tutorial:
option as described in the tutorial: 
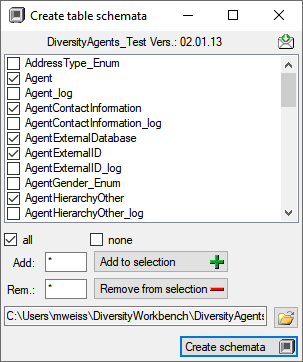
 none buttons resp. the Add to
selection
none buttons resp. the Add to
selection  button
to create the schemata for the selected tables in the predefined
directory.
the
button
to create the schemata for the selected tables in the predefined
directory.
the 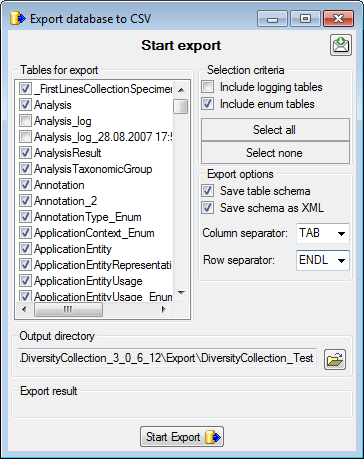
 Backup database from the menu. This
will create a SQL-Server backup on the server where the database is
located. Ensure that there is enough space on the server.
Backup database from the menu. This
will create a SQL-Server backup on the server where the database is
located. Ensure that there is enough space on the server.
 Replicator or Administrator.
Replicator or Administrator.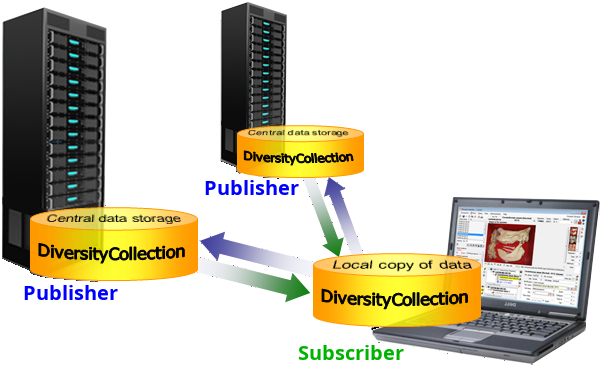
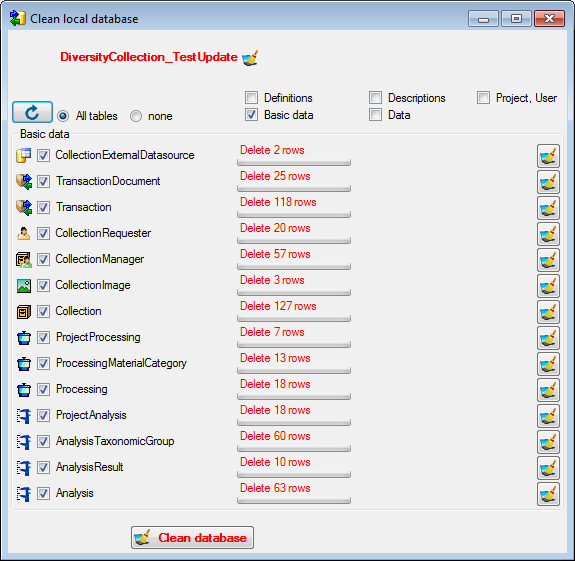
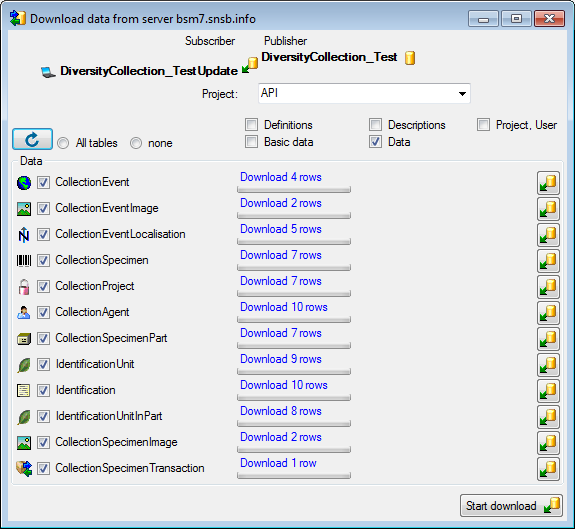
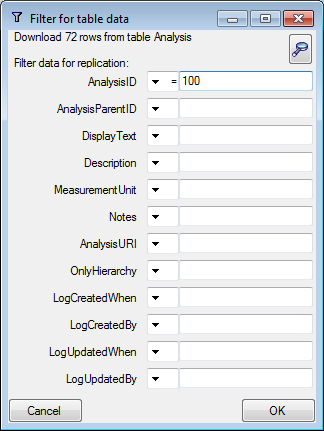
 .
.
 Merge** from the menu ([Publisher] is the
name of the publishing database on the publishing server). As described
for the download, choose the data ranges and click on the
Merge** from the menu ([Publisher] is the
name of the publishing database on the publishing server). As described
for the download, choose the data ranges and click on the
 To fix problems that may interfere with the replication you find some
tools under the menu Data →
To fix problems that may interfere with the replication you find some
tools under the menu Data →
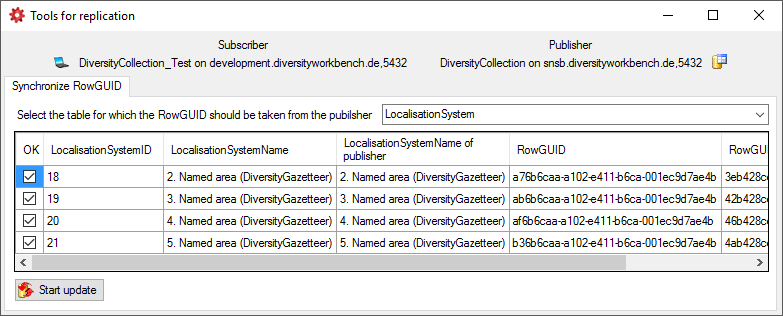
 Start update button to synchronize the
RowGUIDs.
Start update button to synchronize the
RowGUIDs.


 button to open a window as shown below
where you can choose between the two versions of the data as found in
the publisher and the subscriber database.
button to open a window as shown below
where you can choose between the two versions of the data as found in
the publisher and the subscriber database.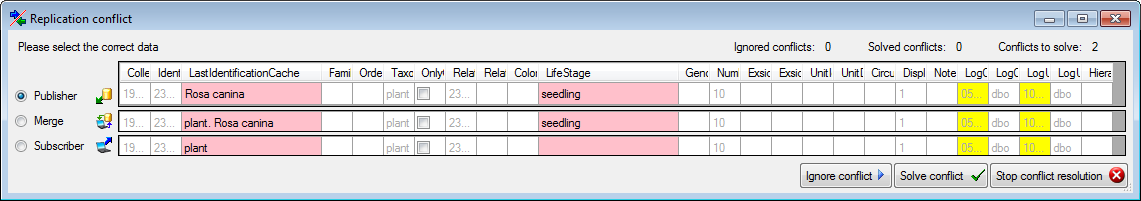
 button. If you can not solve a conflict, use
the Ignore conflict
button. If you can not solve a conflict, use
the Ignore conflict
 buttons respectively.
buttons respectively.
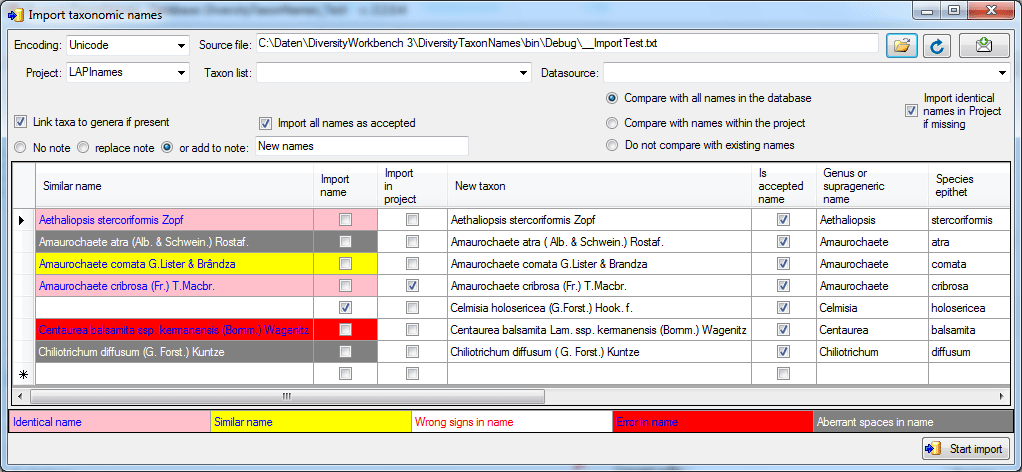



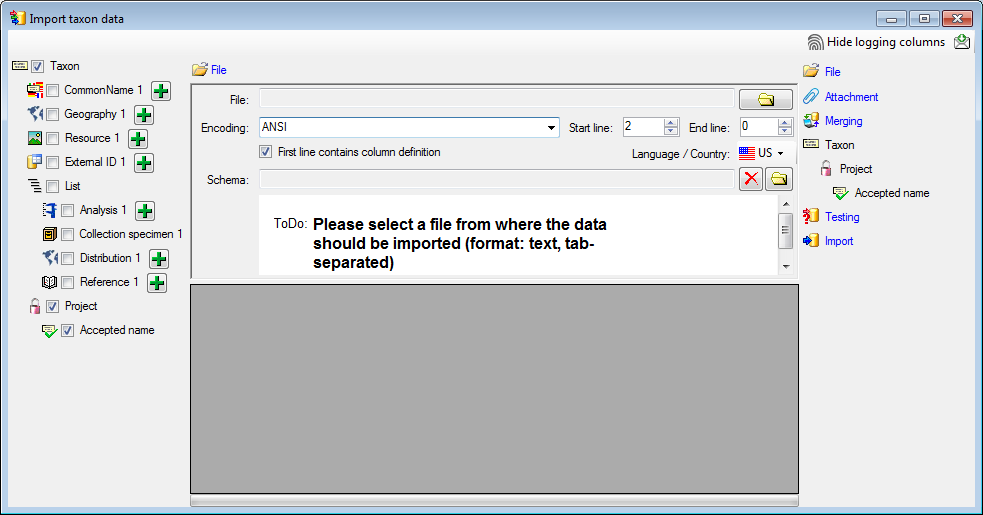
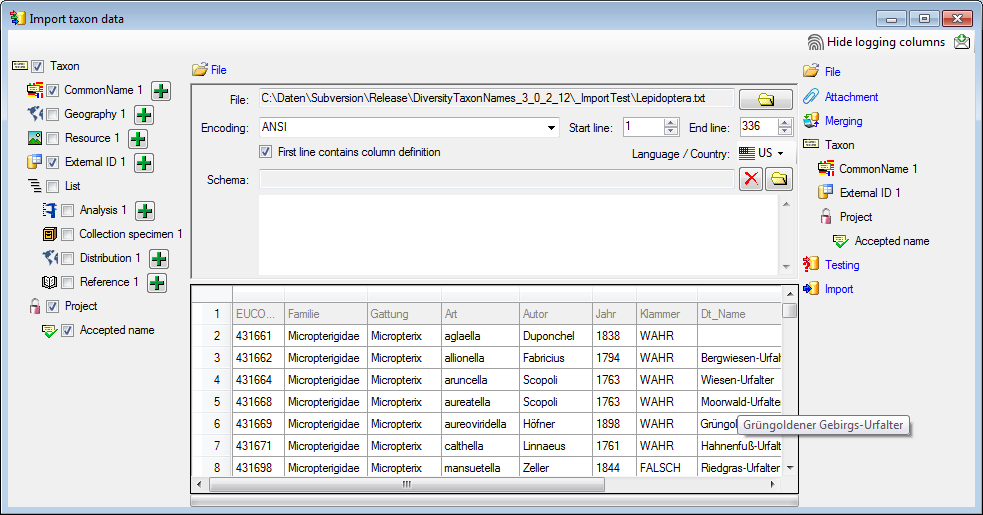
 button in the header line.
This will include a additional substeps for every step containing the
logging columns (see below). If you do not import these data, they will
be automatically filled by default values like the current time and
user.
button in the header line.
This will include a additional substeps for every step containing the
logging columns (see below). If you do not import these data, they will
be automatically filled by default values like the current time and
user.
 Attach them to data in the database. Select the import step
Attach them to data in the database. Select the import step
 Import as
new data or one of the columns the attachment columns offered like
SeriesCode in the table Series in the example below.
Import as
new data or one of the columns the attachment columns offered like
SeriesCode in the table Series in the example below.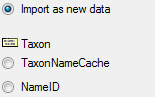

 Insert,
Insert,  Update and
Update and
 Key columns (see below). If no matching data are
found in the database, the data from the file will be imported,
otherwise the data will be updated..
Key columns (see below). If no matching data are
found in the database, the data from the file will be imported,
otherwise the data will be updated..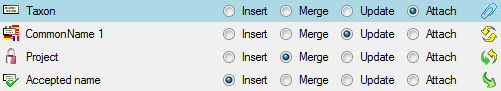
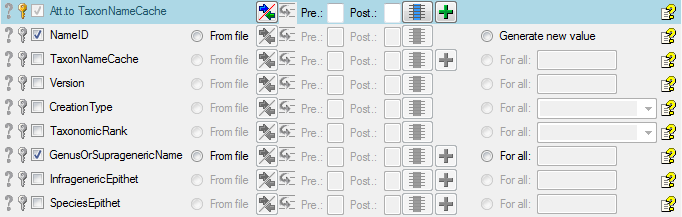
 =
If data will be imported depends on the content of decisive colums, so
at least one must be selected.
=
If data will be imported depends on the content of decisive colums, so
at least one must be selected. =
The position in the file must be given if the data for a column should
be taken from the file.
=
The position in the file must be given if the data for a column should
be taken from the file. = You have to enter a
value used for all datasets
= You have to enter a
value used for all datasets Testing step. You can use a certain line in
the file for you test and then click on the Test data in line:
button. If there are still unmet requirements, these will be listed in a
window as shown below.
Testing step. You can use a certain line in
the file for you test and then click on the Test data in line:
button. If there are still unmet requirements, these will be listed in a
window as shown below.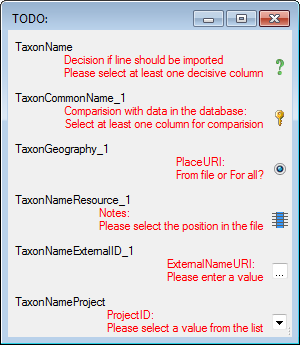
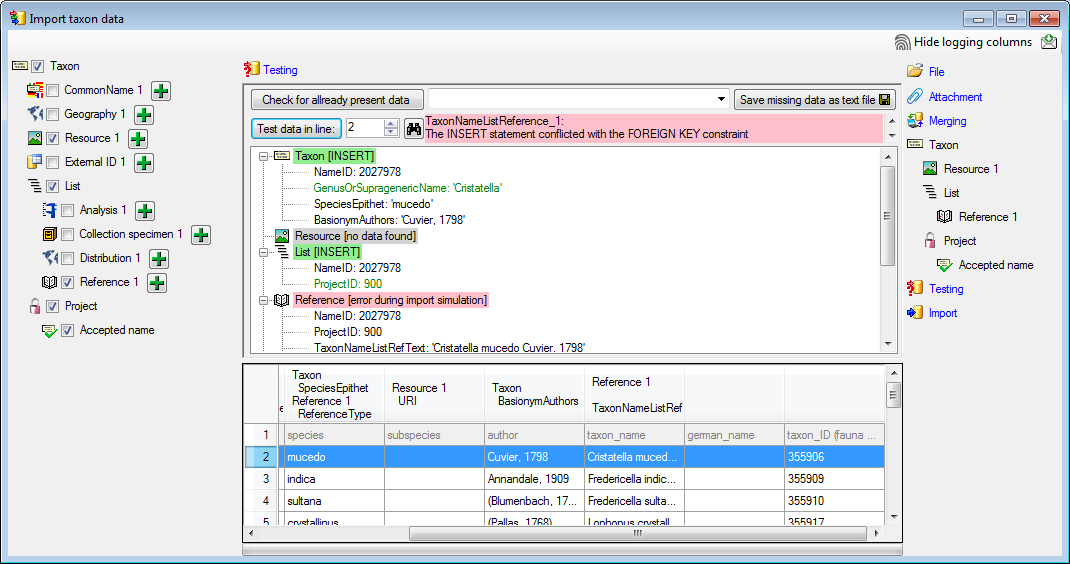
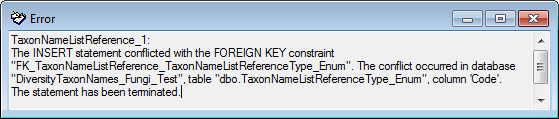
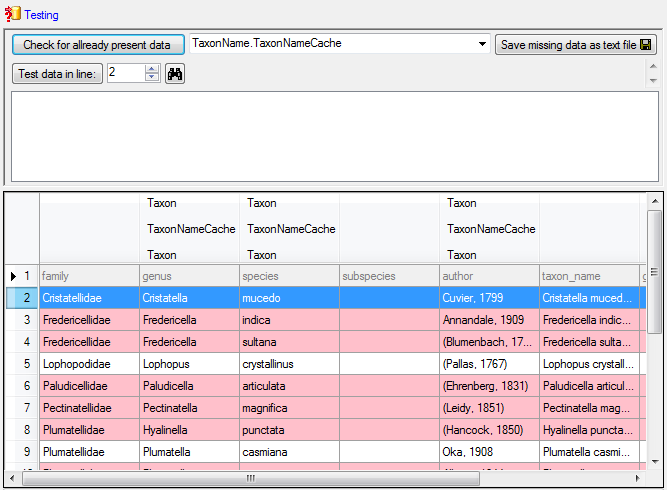
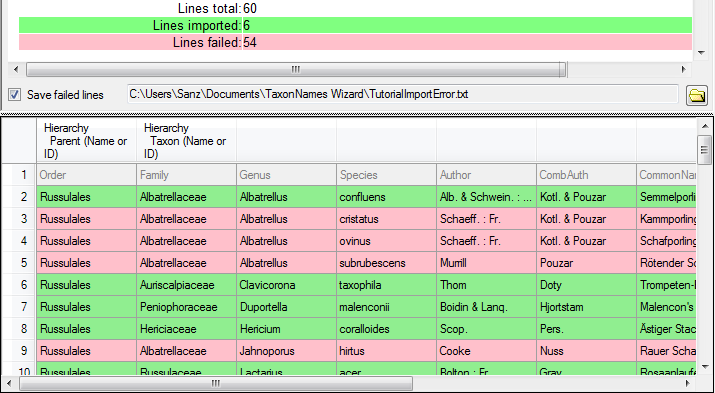
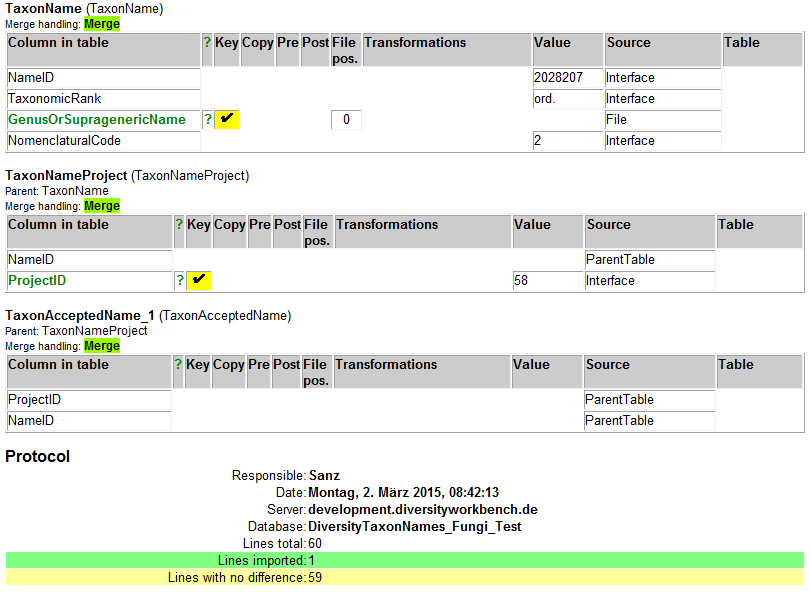

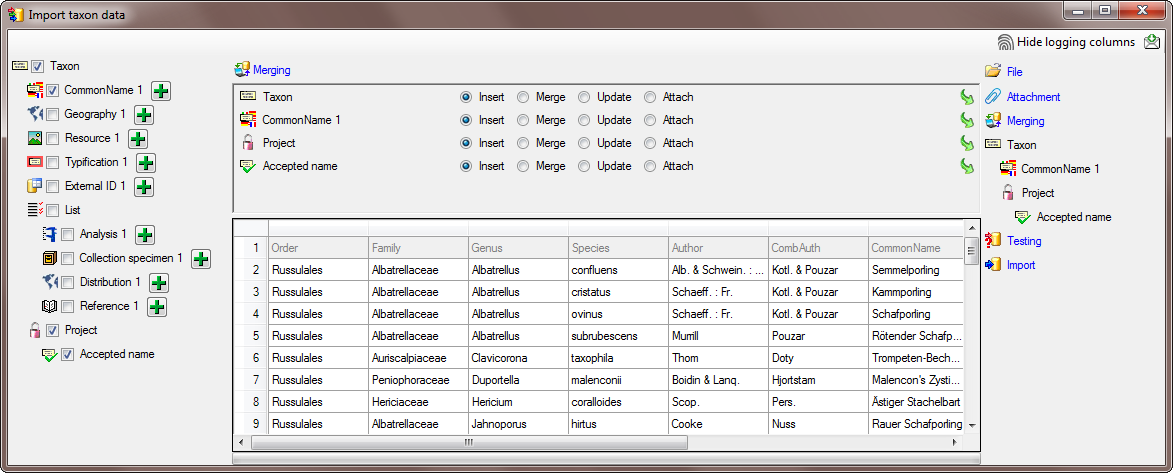
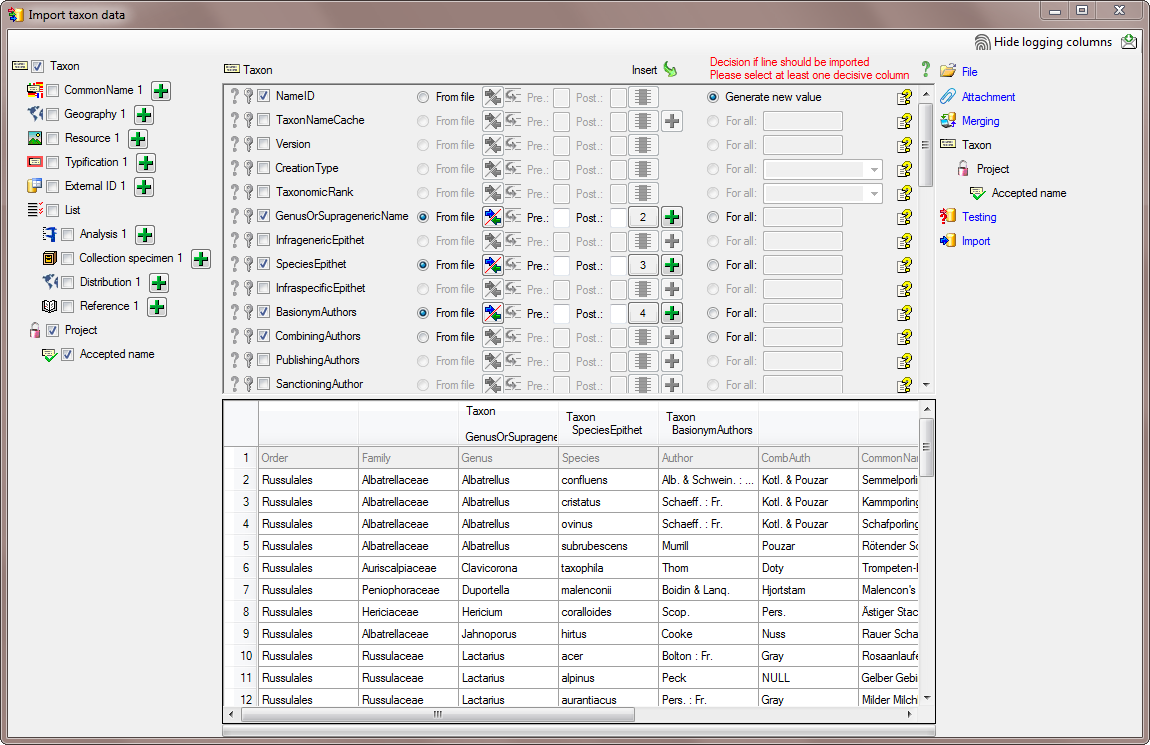
 button) and press the
button) and press the 
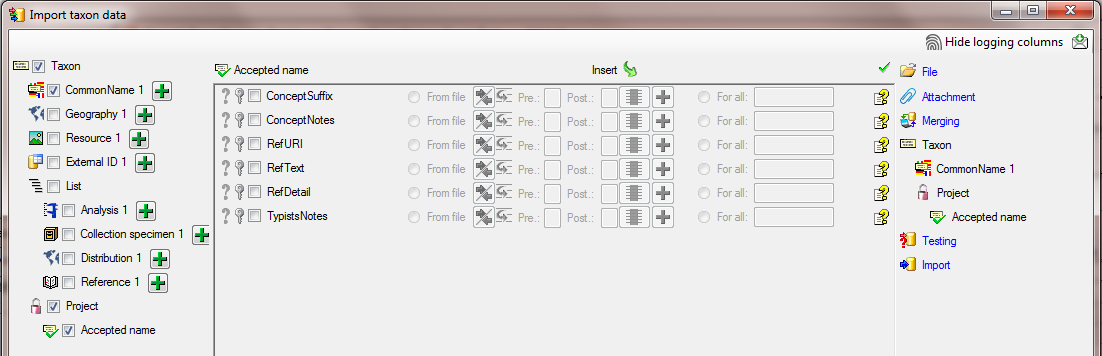
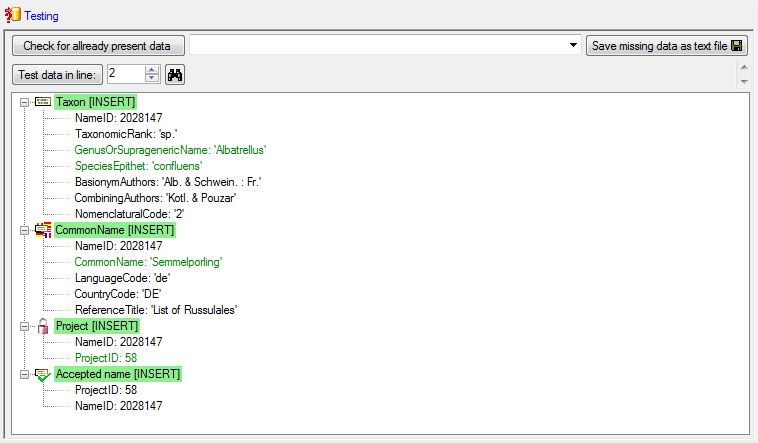
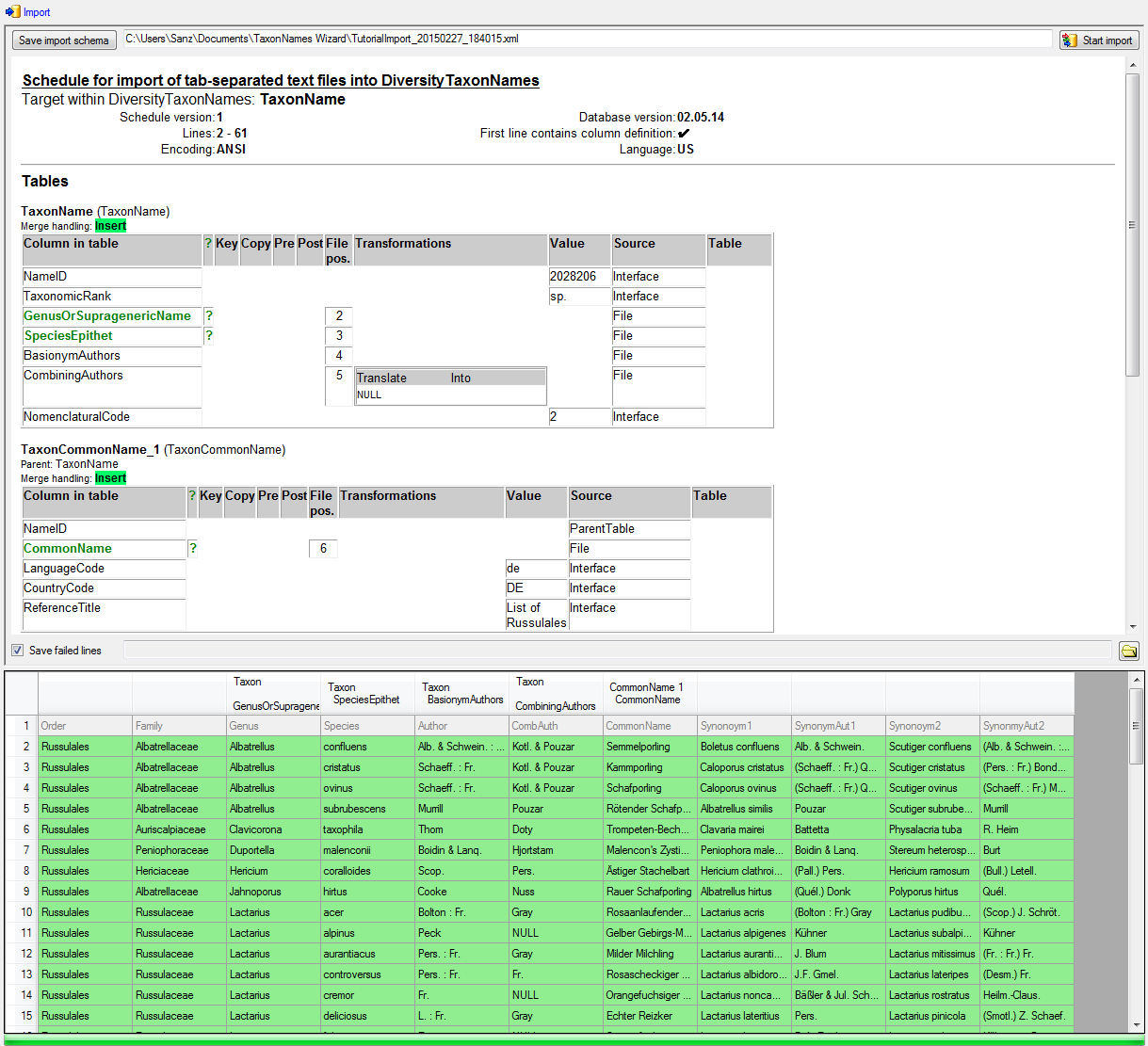
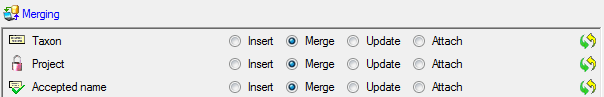
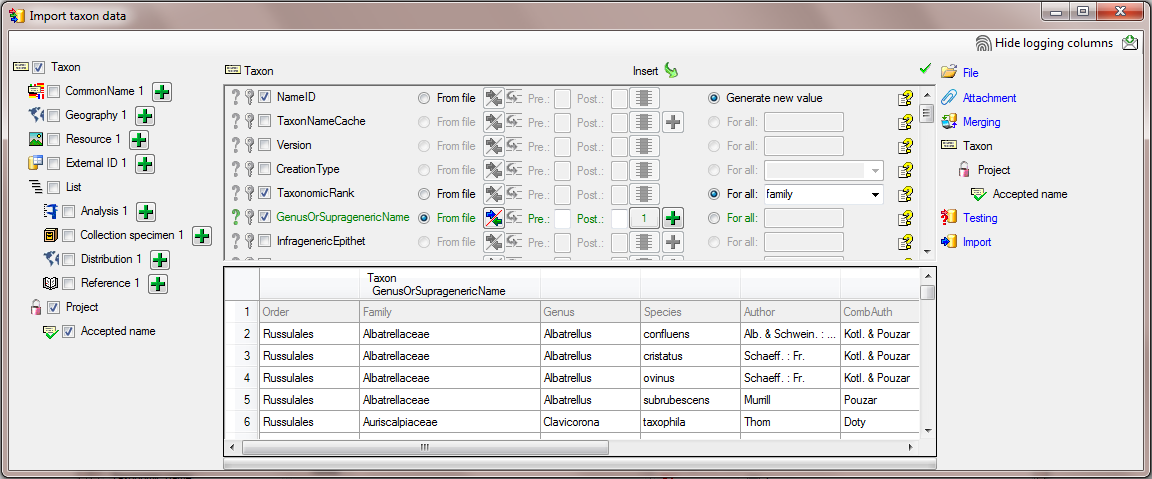
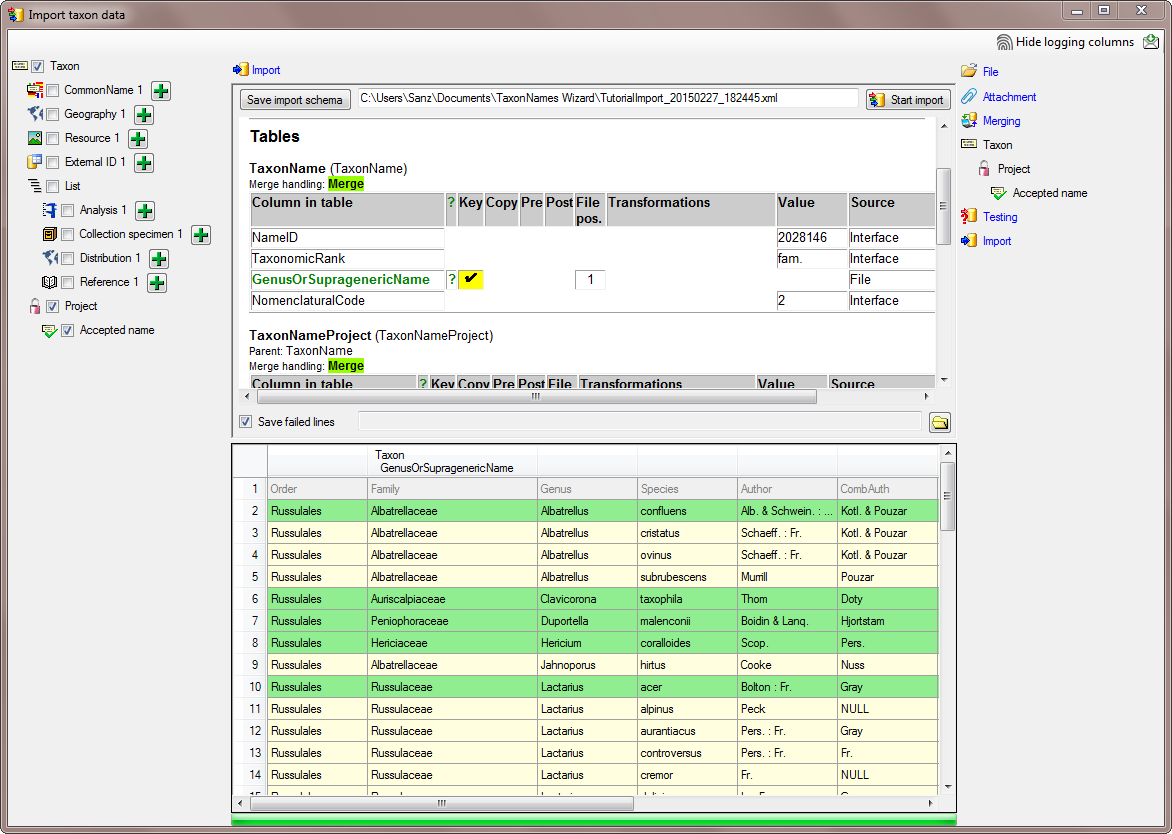
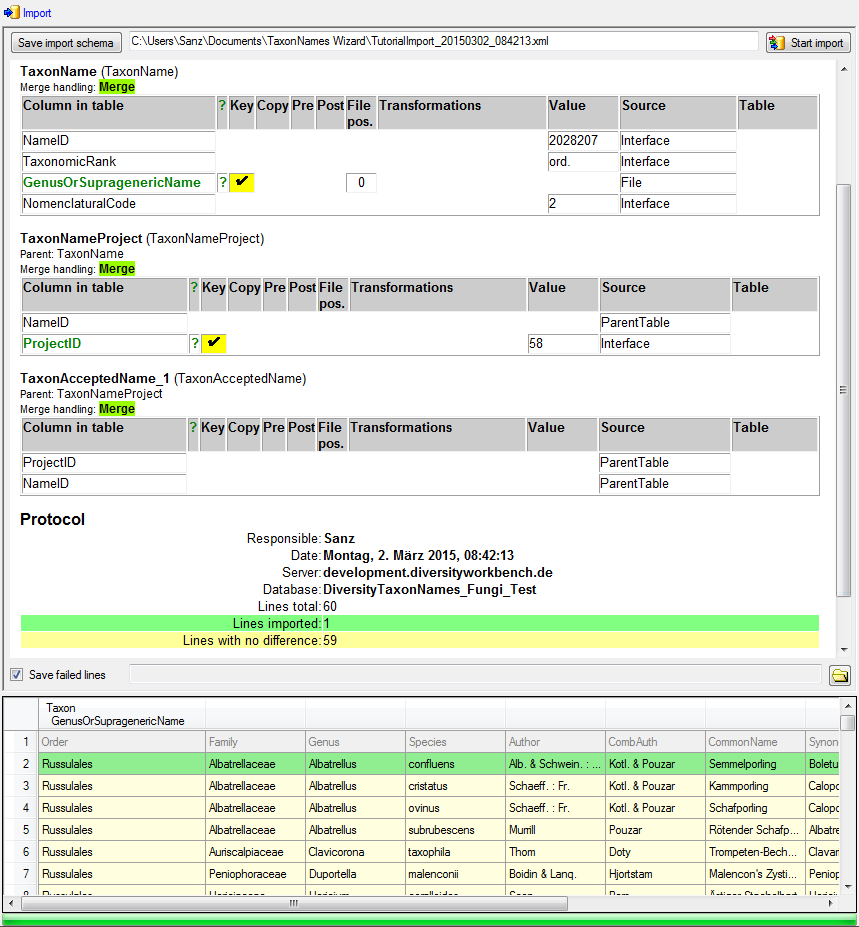
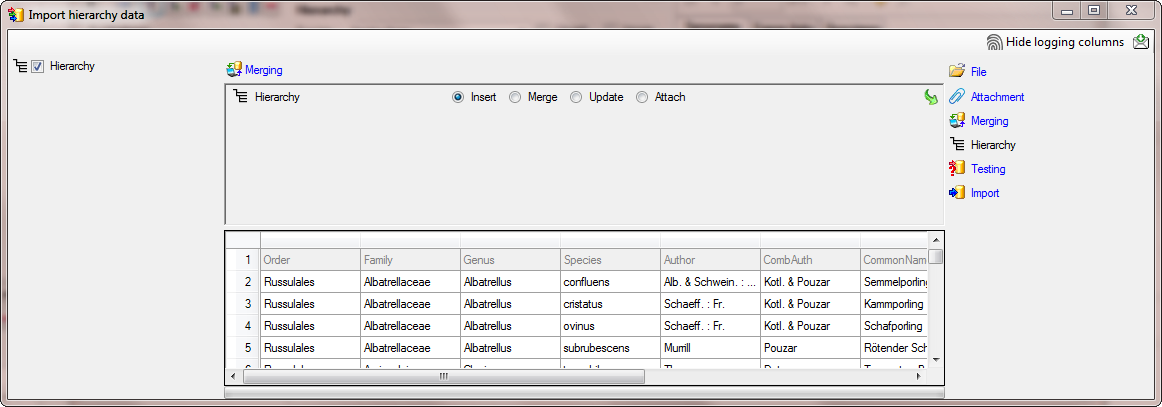
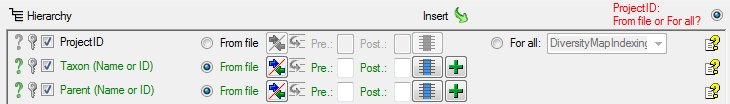
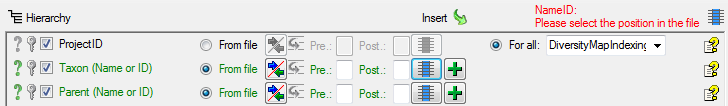
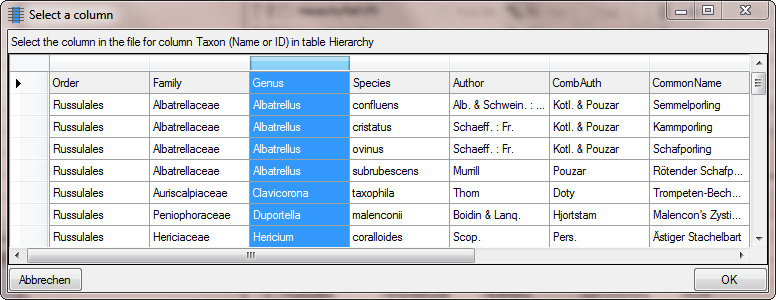
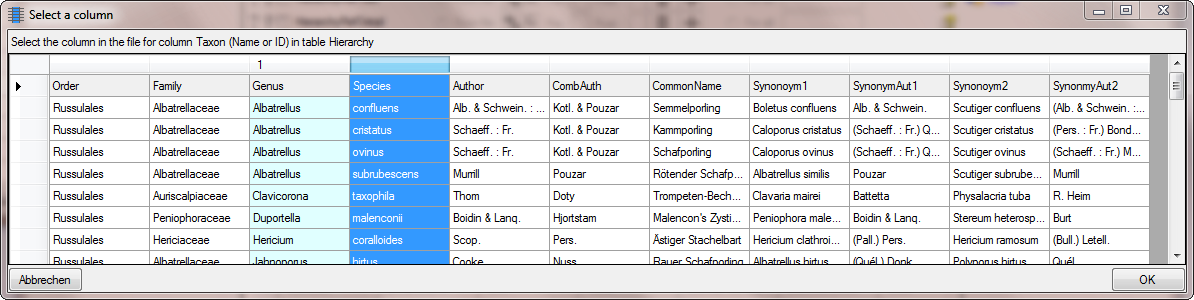
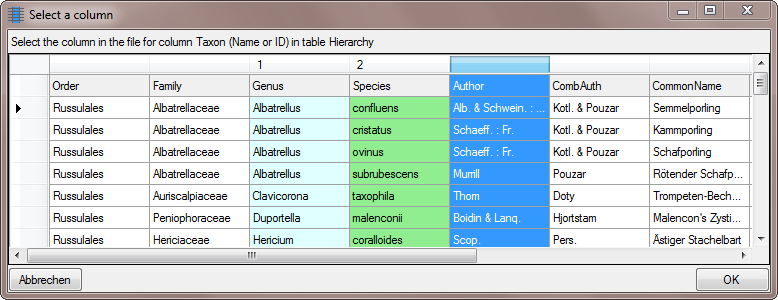
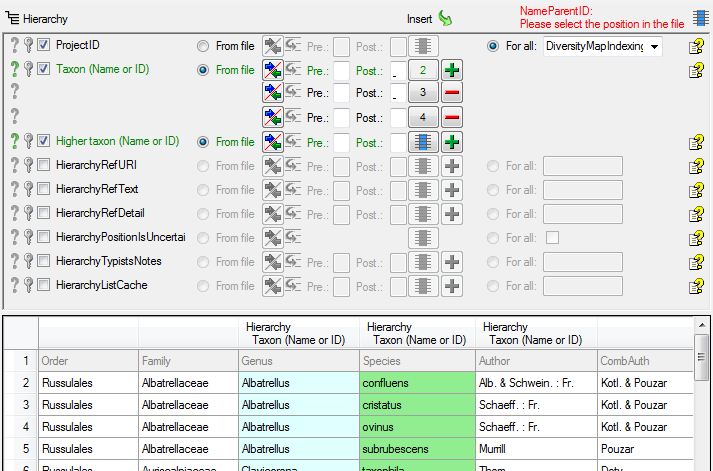
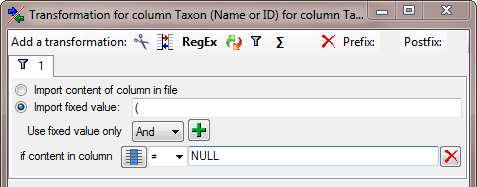
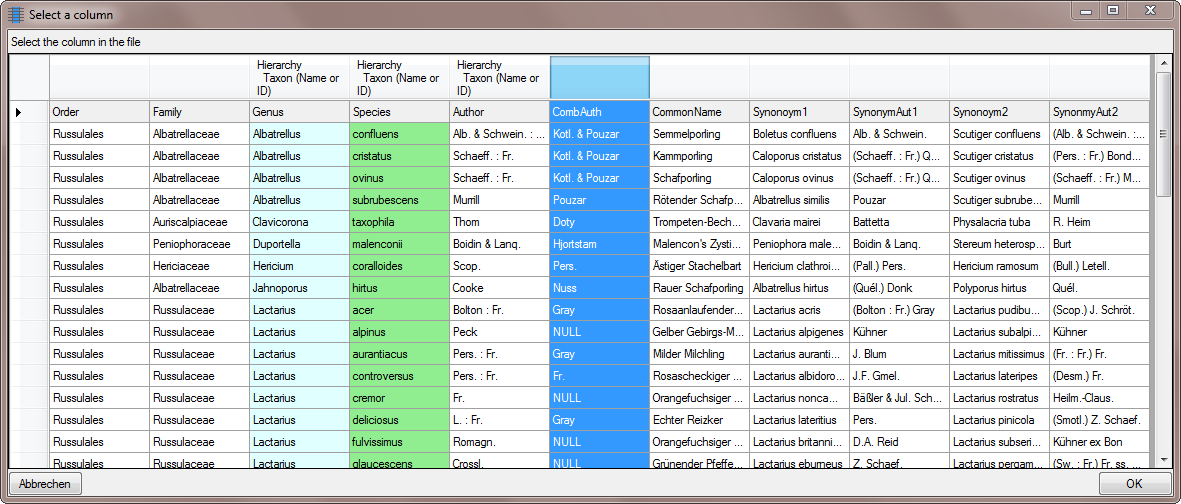
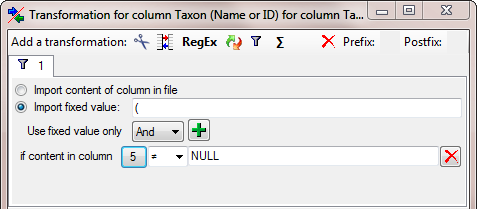
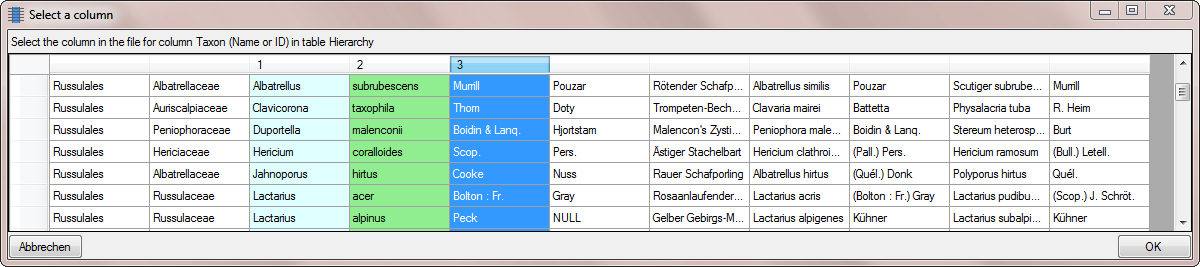
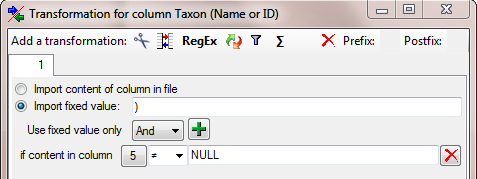
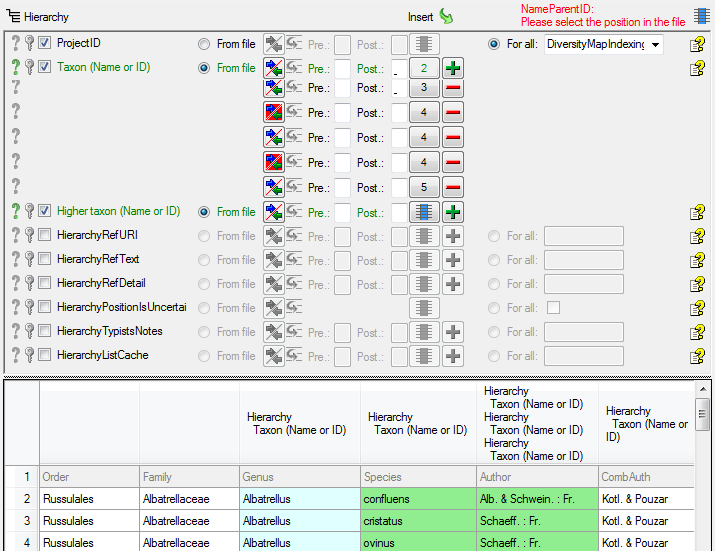
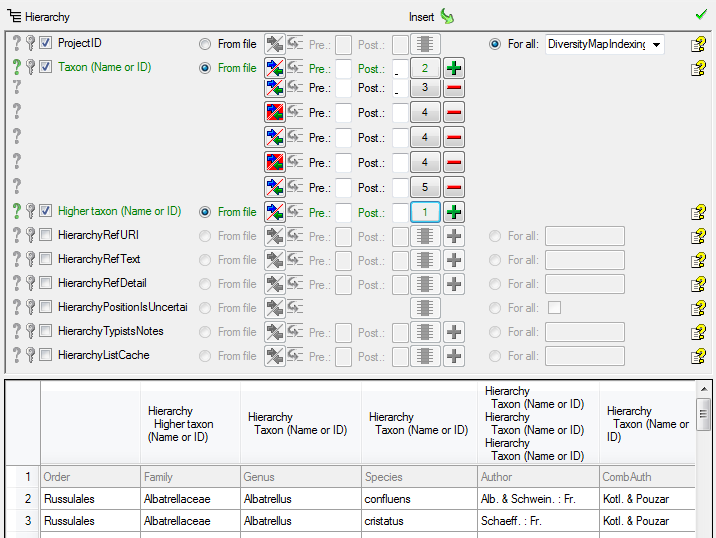
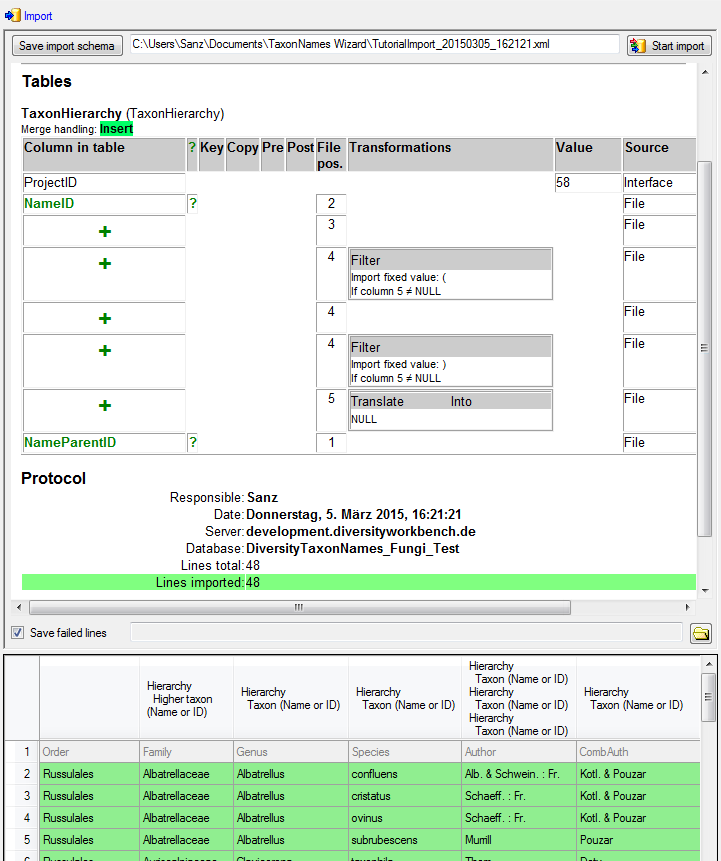
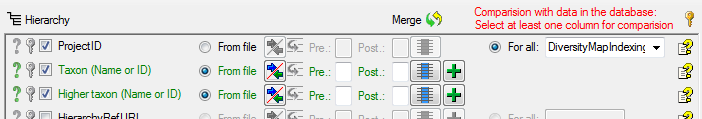
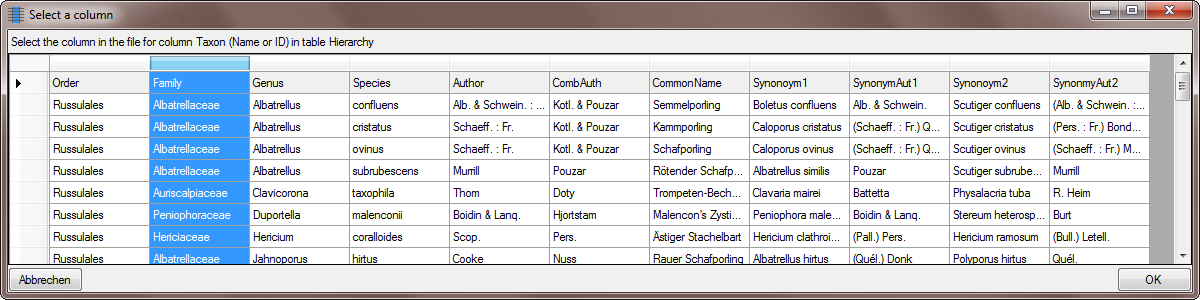
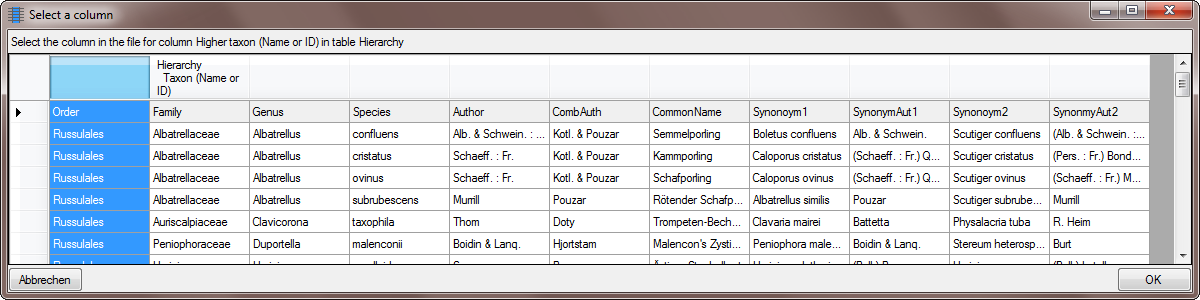
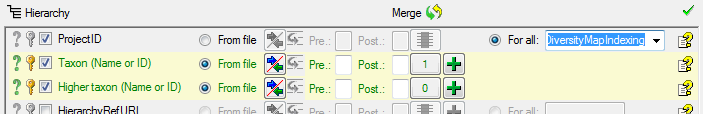
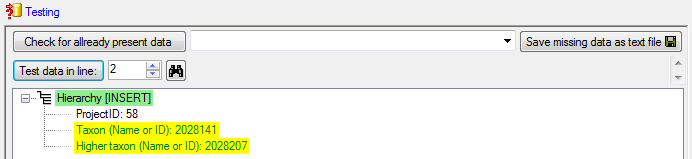
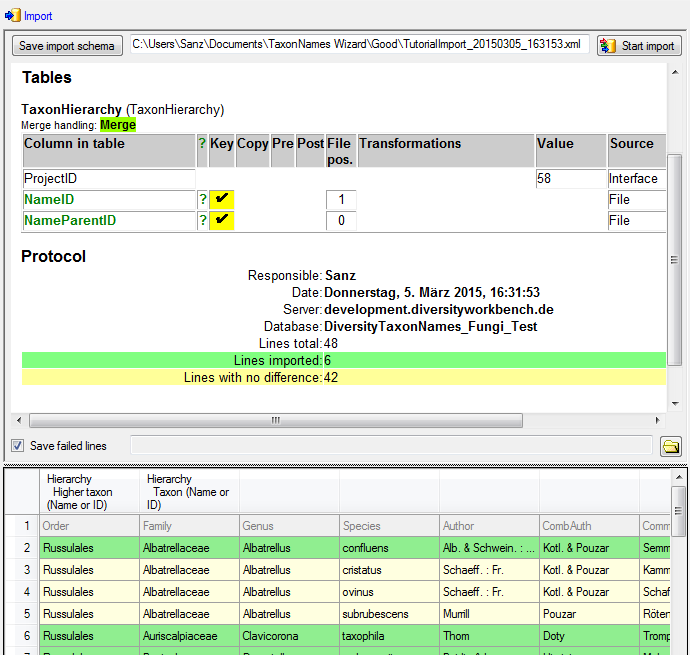
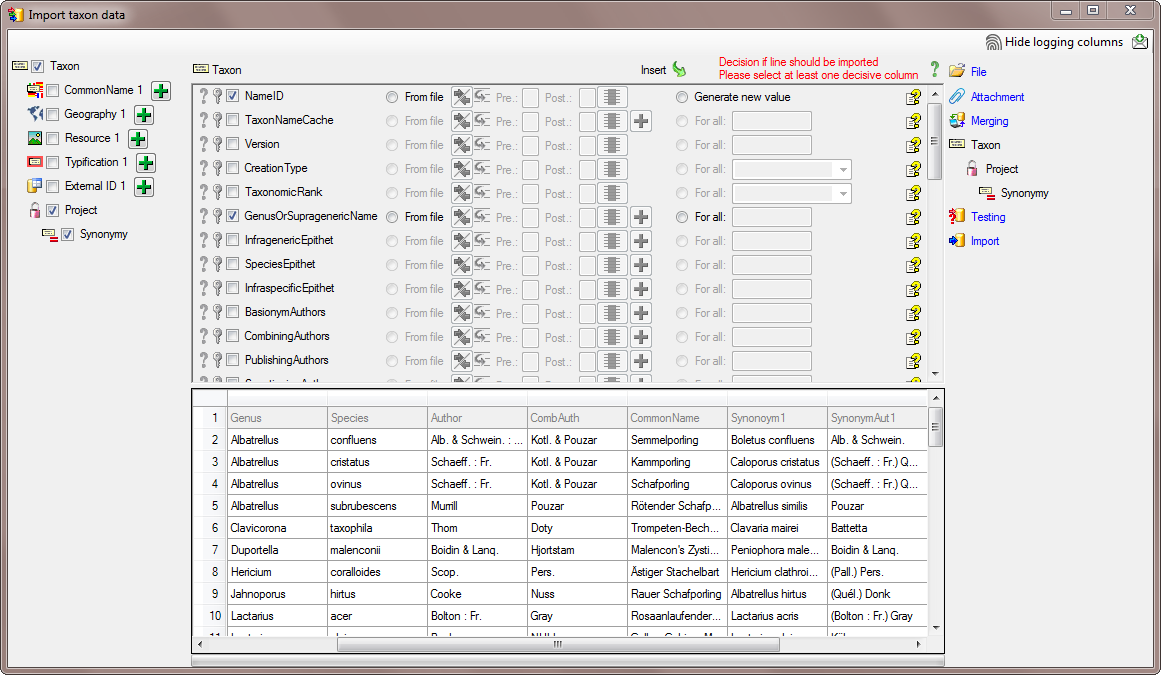
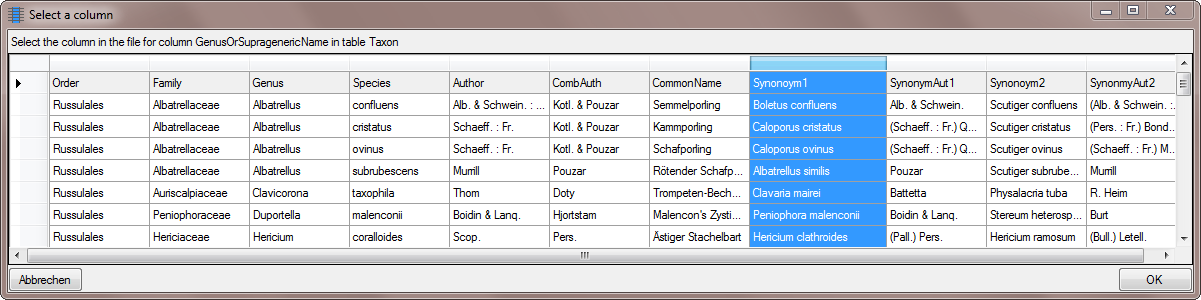
 cutting transformation.
Enter a space as Splitter. A Test of the transformation should produce
the result as shown below.
cutting transformation.
Enter a space as Splitter. A Test of the transformation should produce
the result as shown below.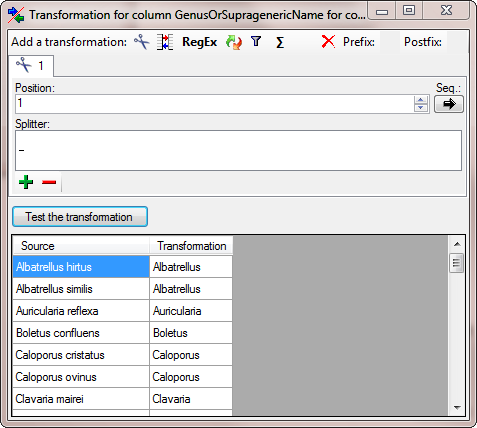
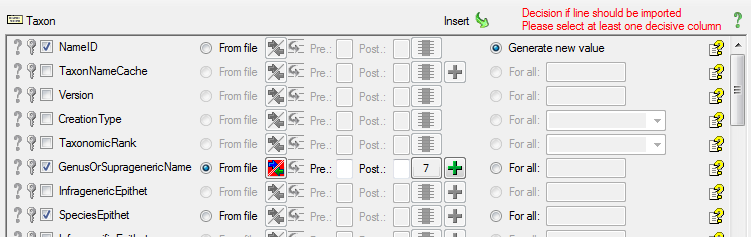
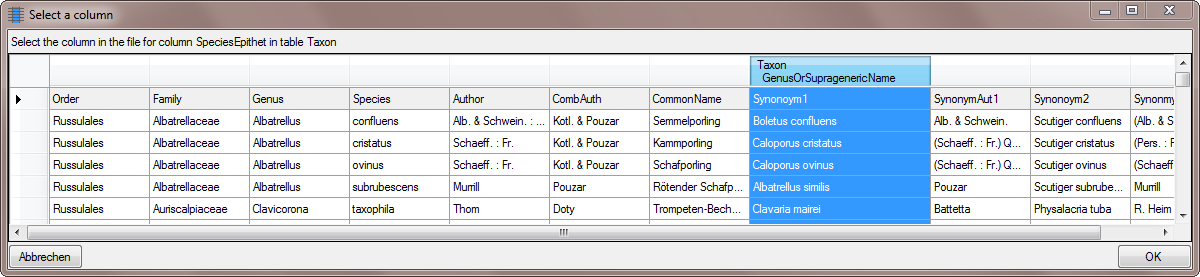
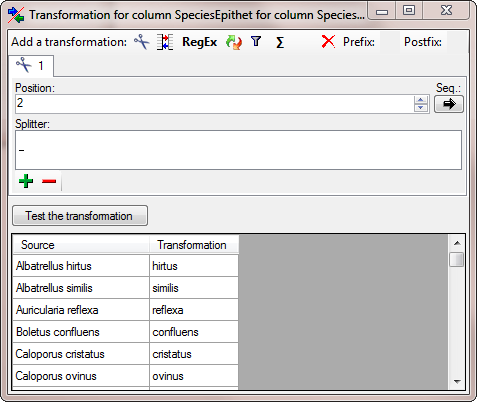
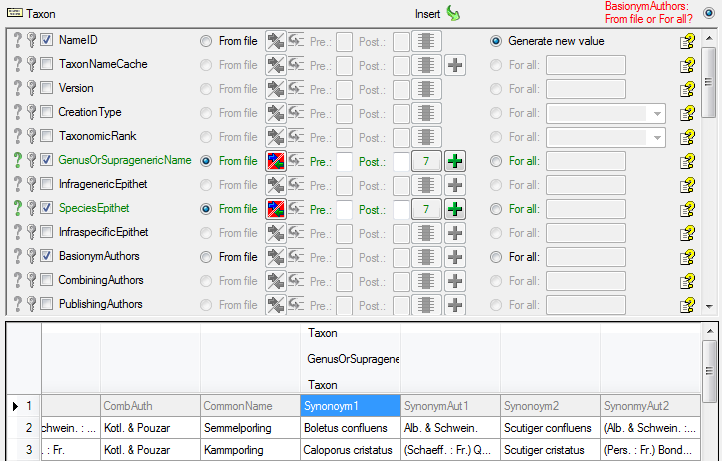
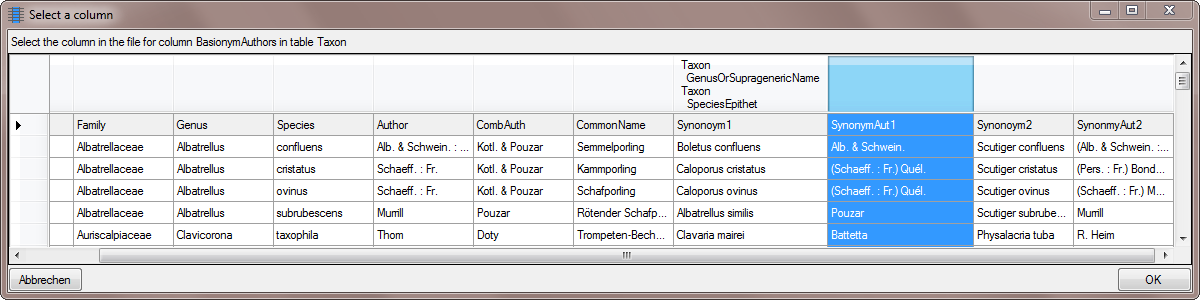
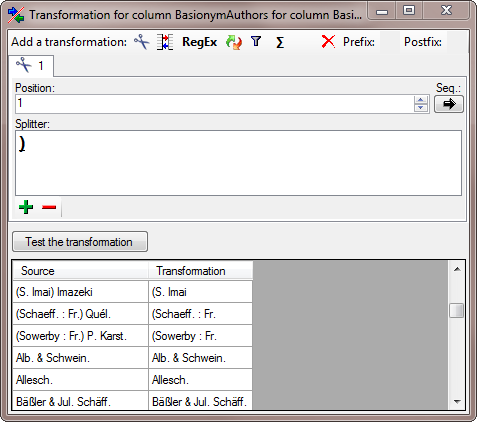
 Replace transformation to get
rid of the leeding bracket (see below).
Replace transformation to get
rid of the leeding bracket (see below).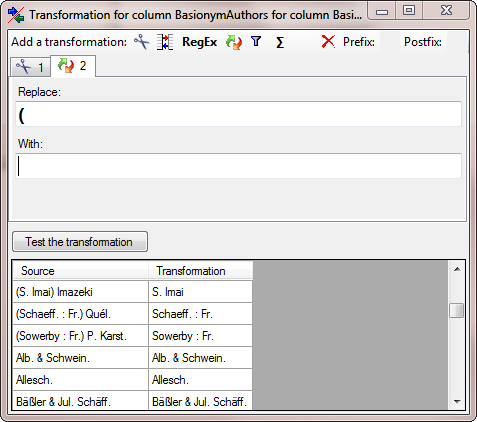
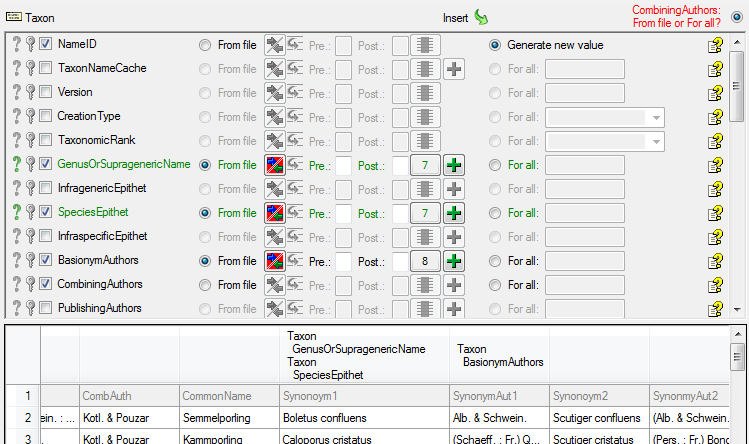
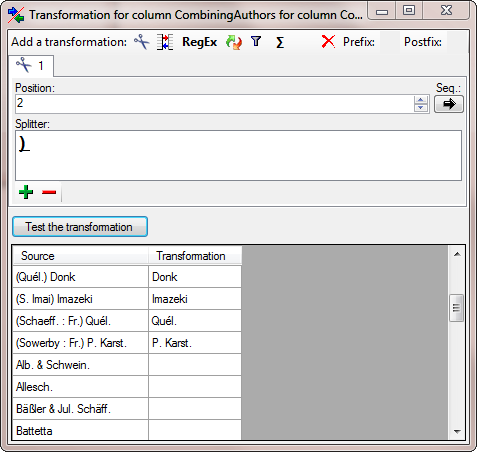
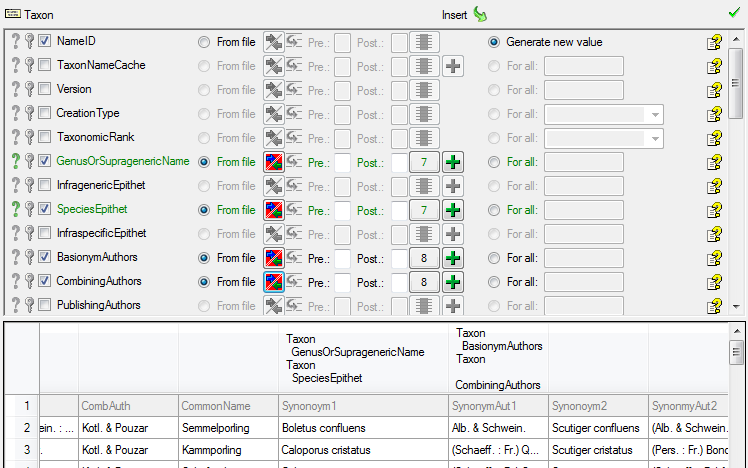
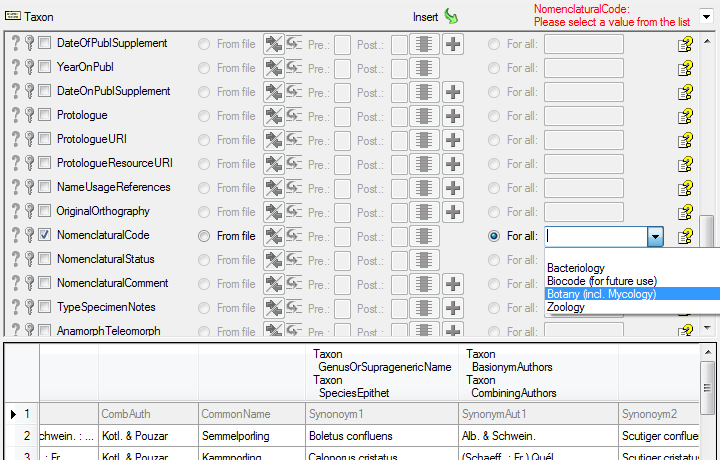

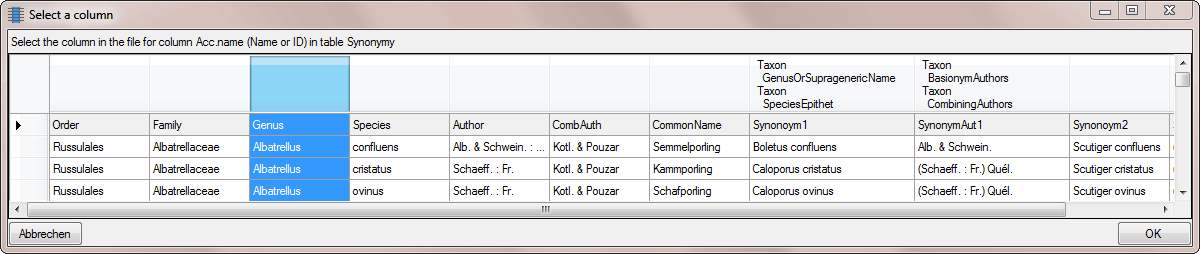
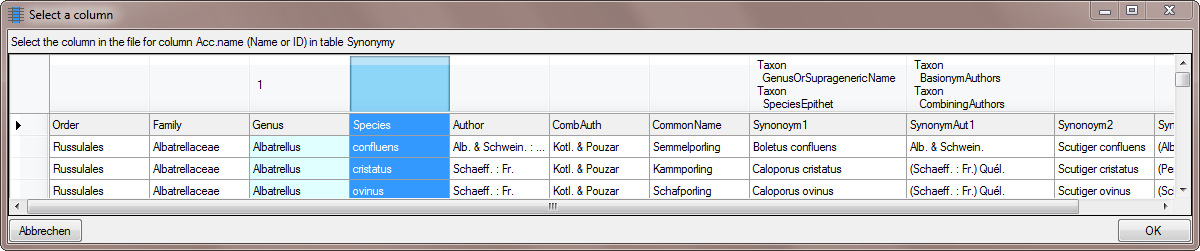
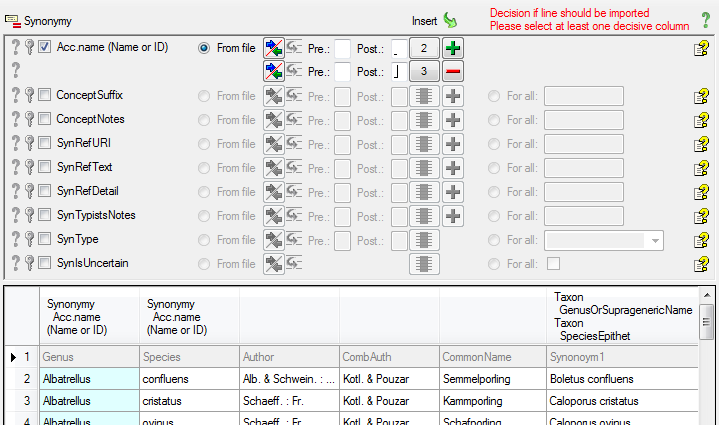
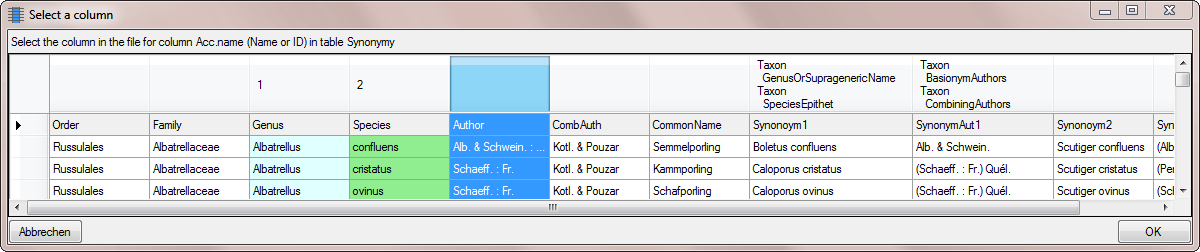
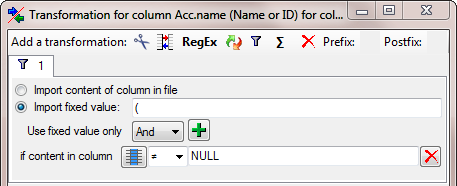
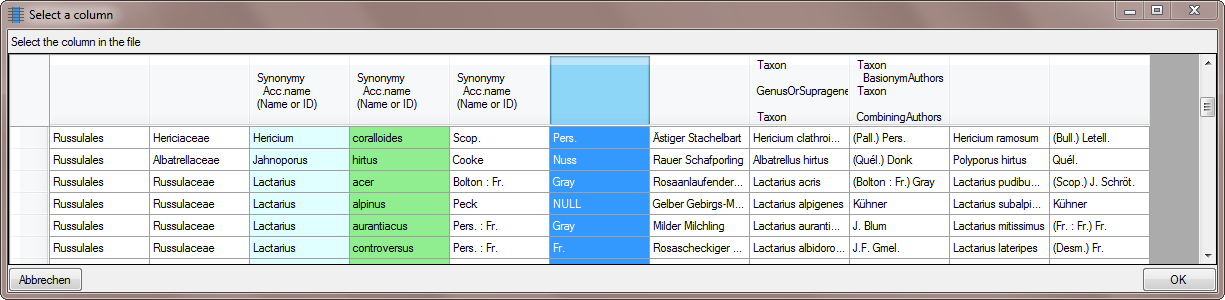
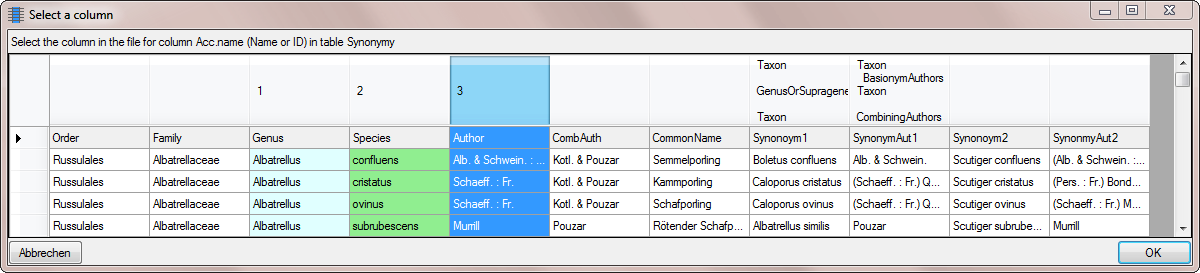
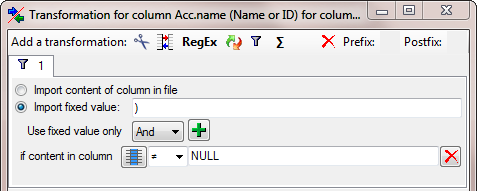
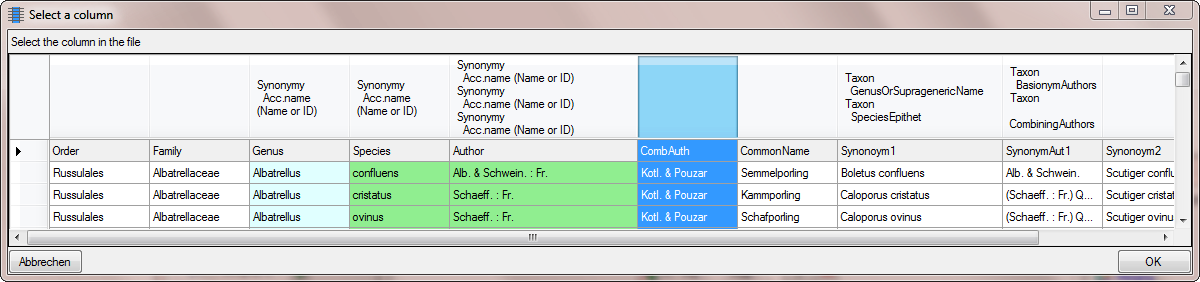
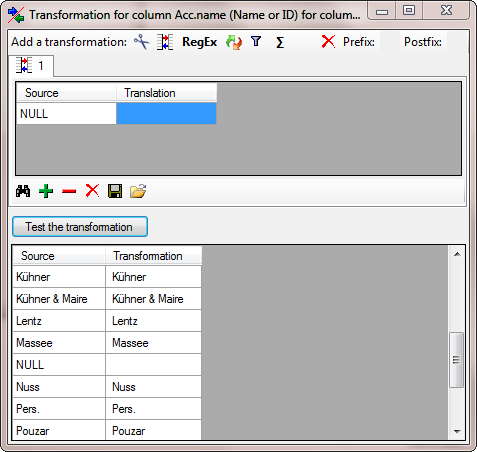
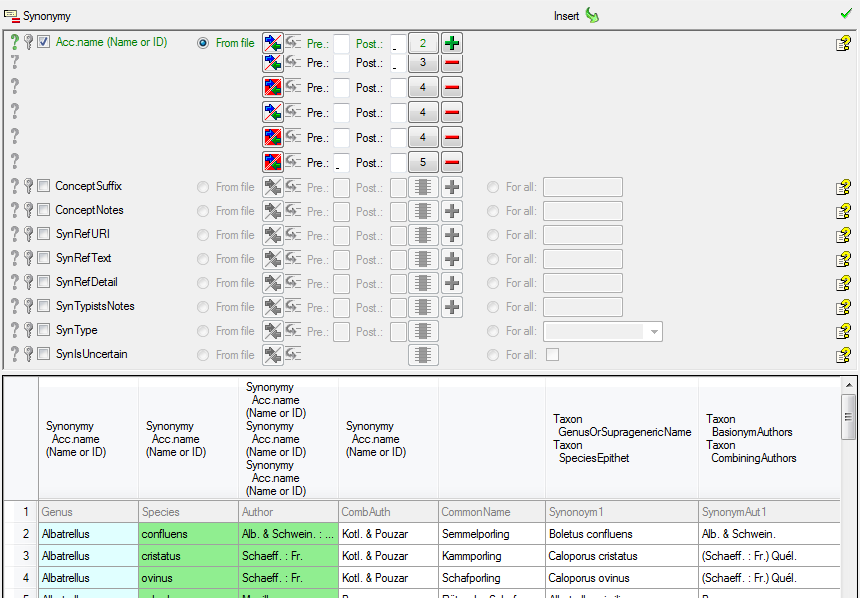
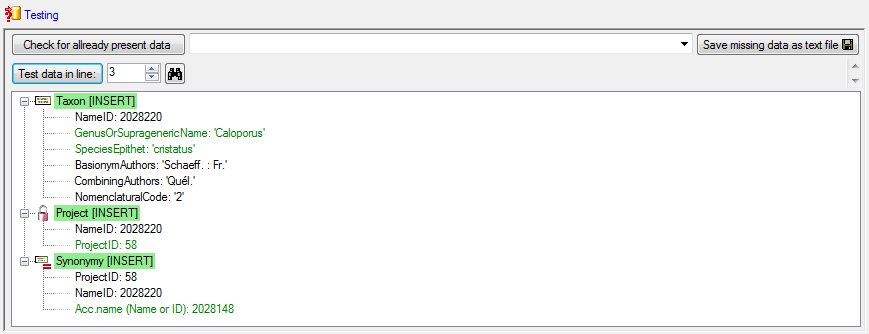
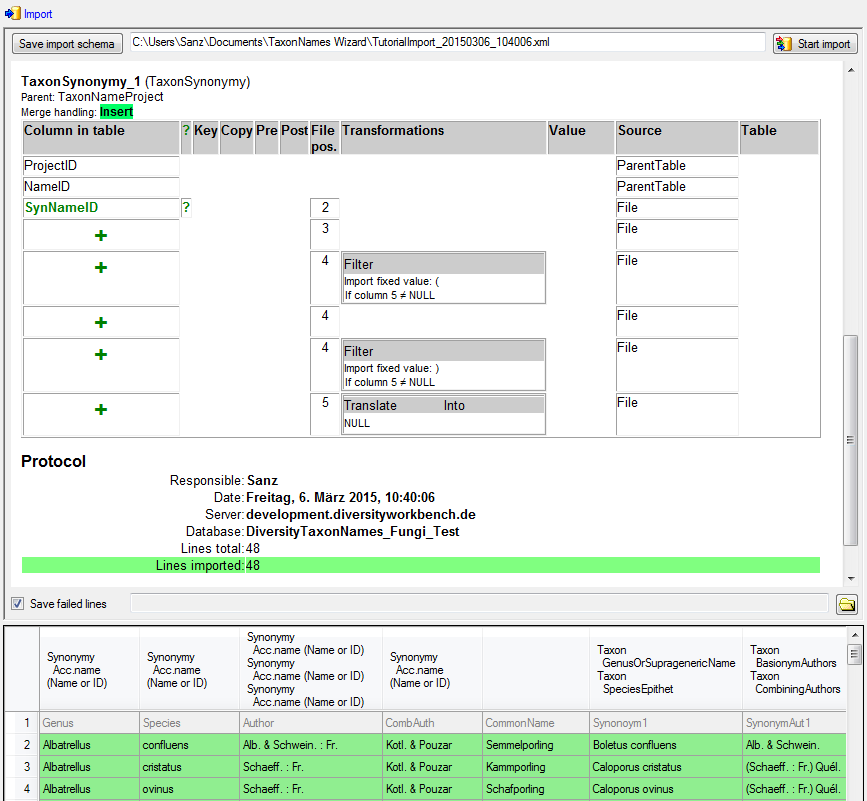
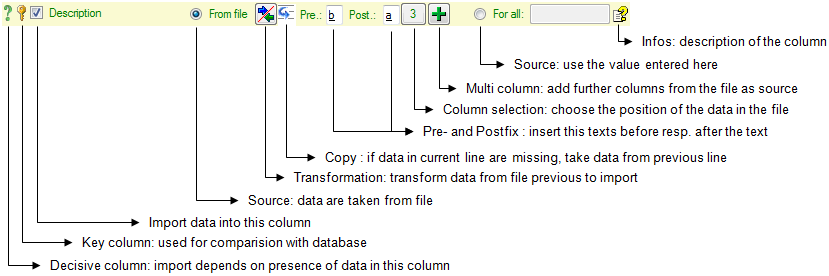

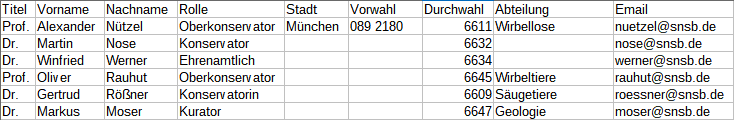
 Copy line option to fill in
missing data as shown below where the blue
values are copied into empty fields during the
import. Click on the
Copy line option to fill in
missing data as shown below where the blue
values are copied into empty fields during the
import. Click on the 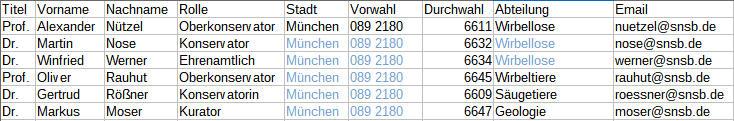
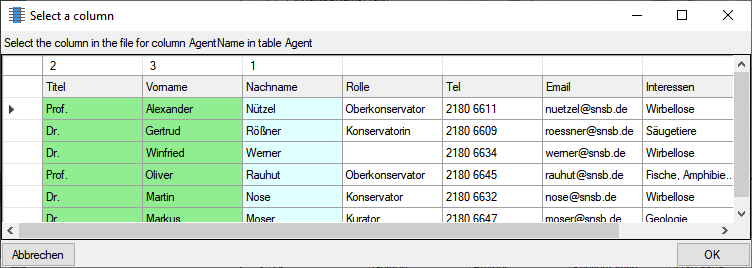
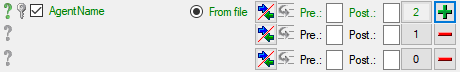
 Seq starting at the first position and
Seq starting at the first position and
 starting at the last position. Click on
the button Test the transformation to see the result of your
transformation.
starting at the last position. Click on
the button Test the transformation to see the result of your
transformation.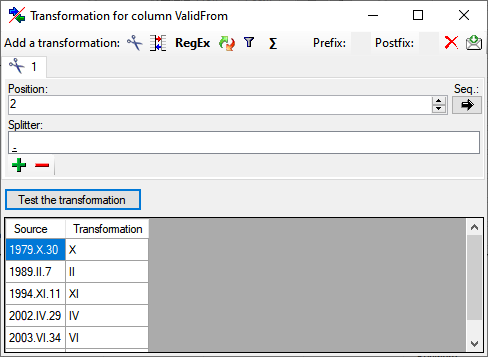
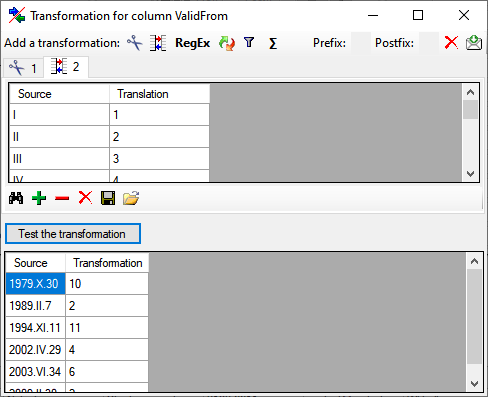
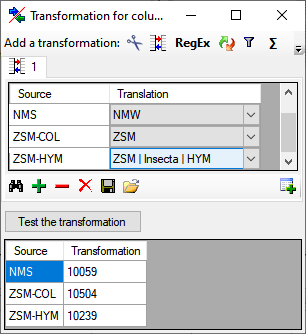
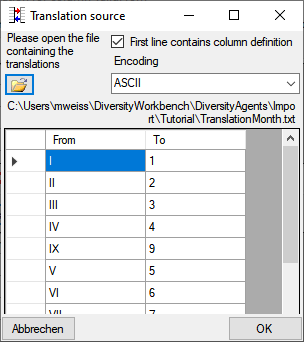
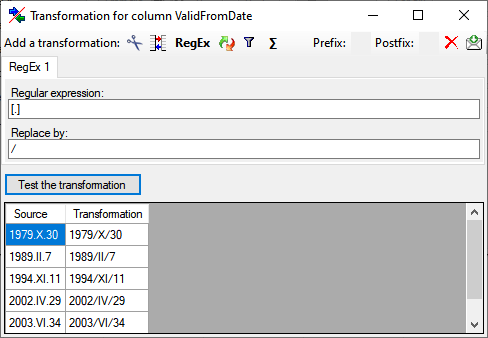
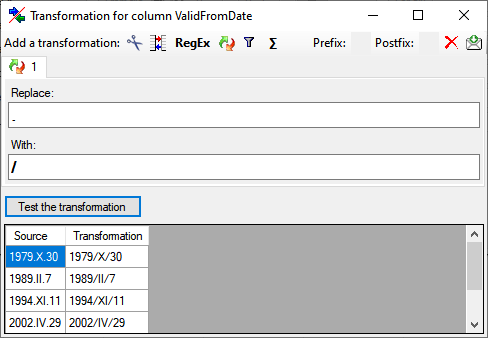
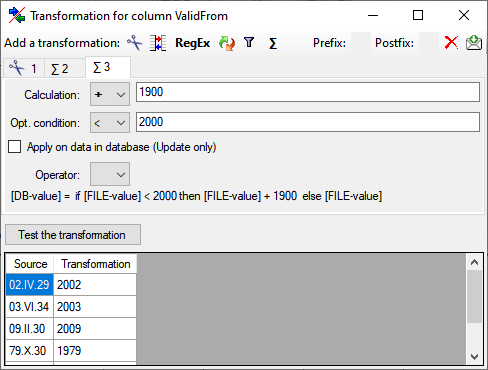
 Several parallel tables according to
selected data
Several parallel tables according to
selected data Dependent table
Dependent table or
descending
or
descending  .
. on the left side of the column that will change to
on the left side of the column that will change to  for fused columns. To remove a file column, use the
for fused columns. To remove a file column, use the  This filter in contrast to the filter above strictly applies to the row
according to the sequence of the data. For an explanation see a short
tutorial
This filter in contrast to the filter above strictly applies to the row
according to the sequence of the data. For an explanation see a short
tutorial
 To test the export choose the
To test the export choose the  To export your data into a
To export your data into a 
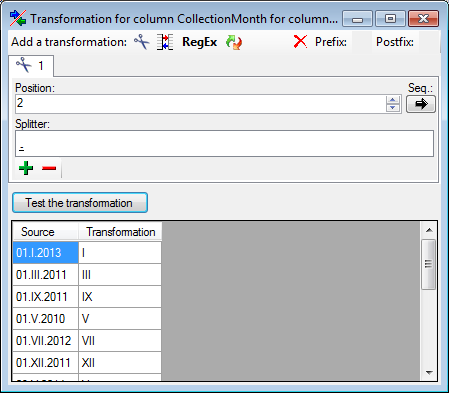
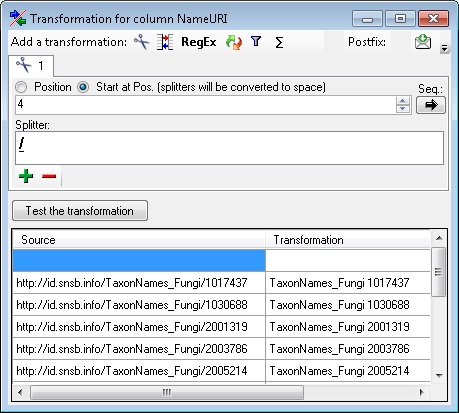
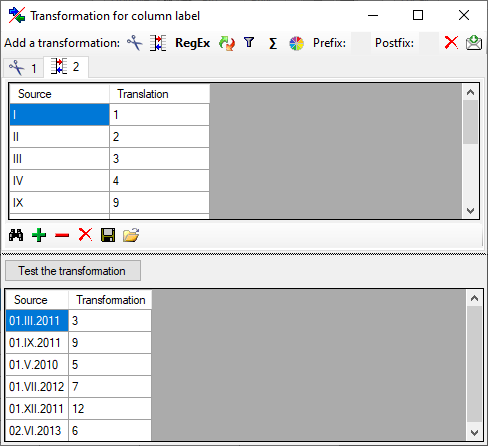
 First line contains column definition. Click OK to use the values
from the file for the translation.
First line contains column definition. Click OK to use the values
from the file for the translation.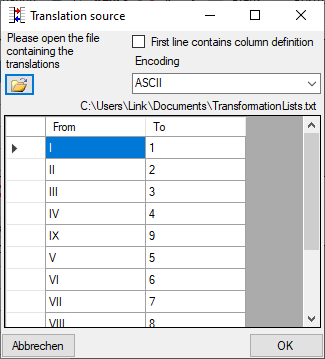
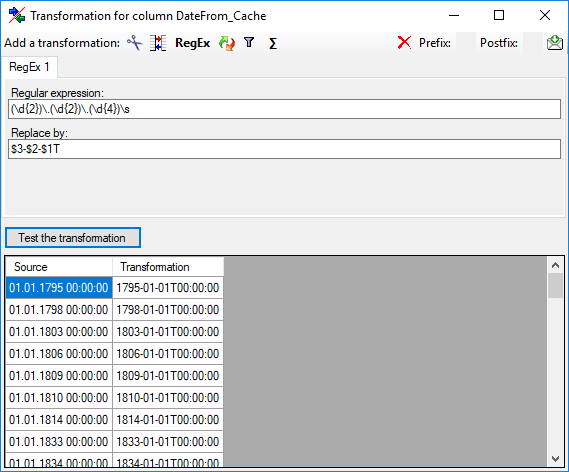
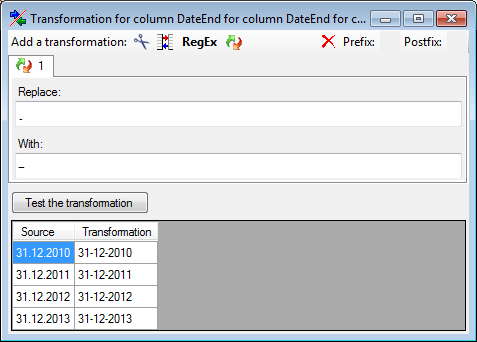
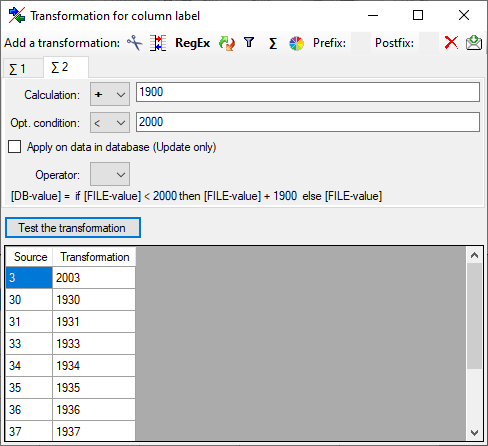
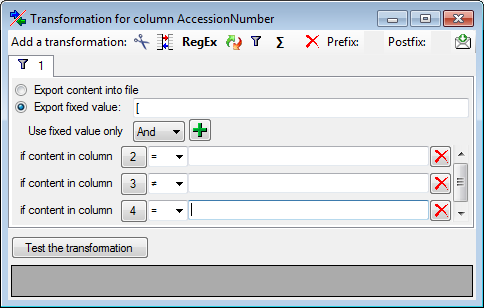
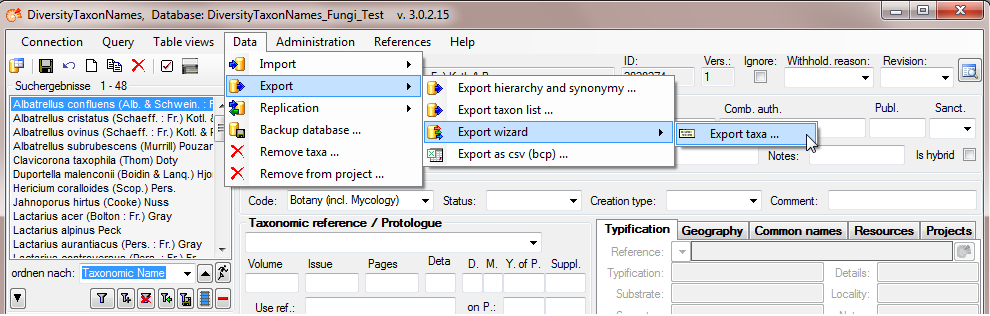
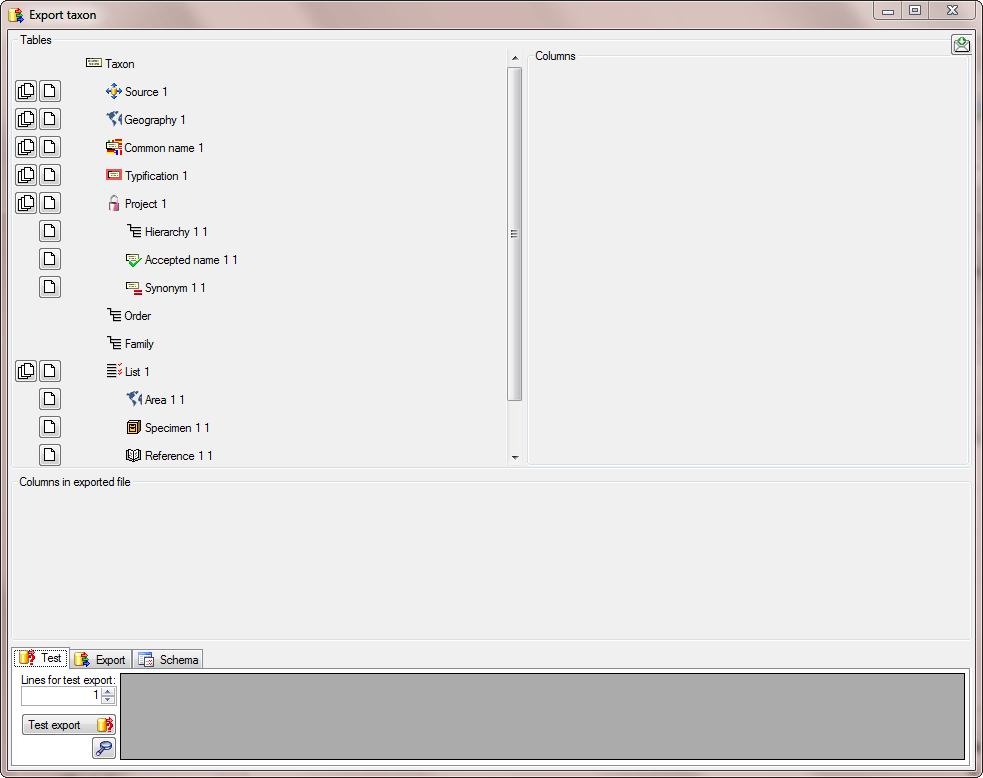
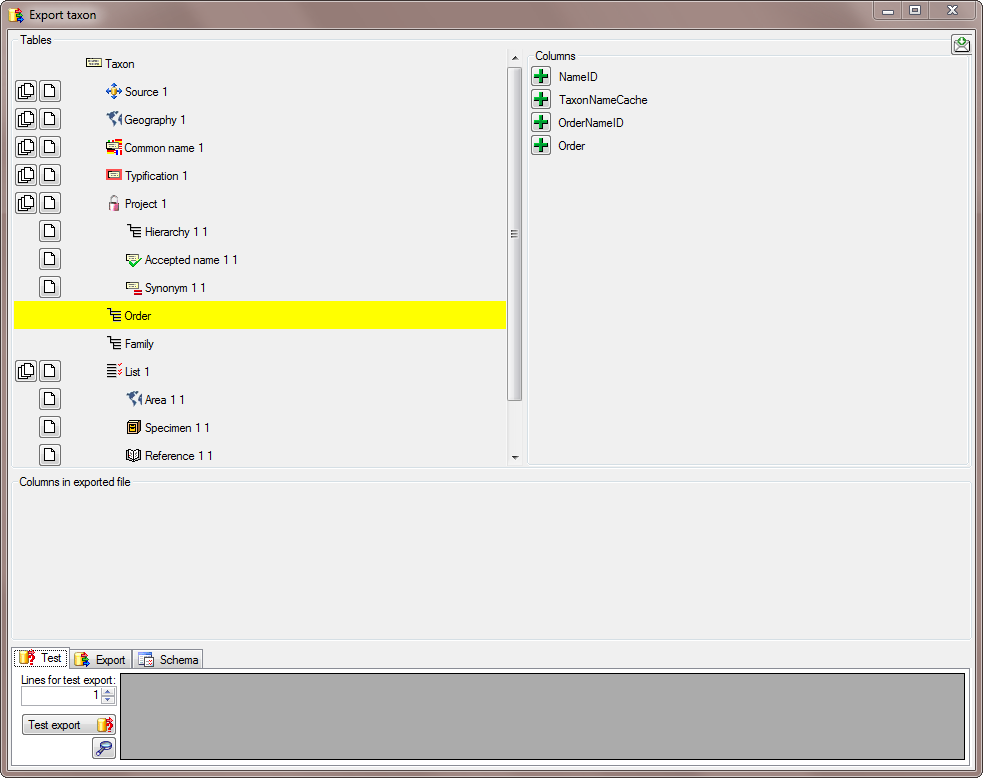
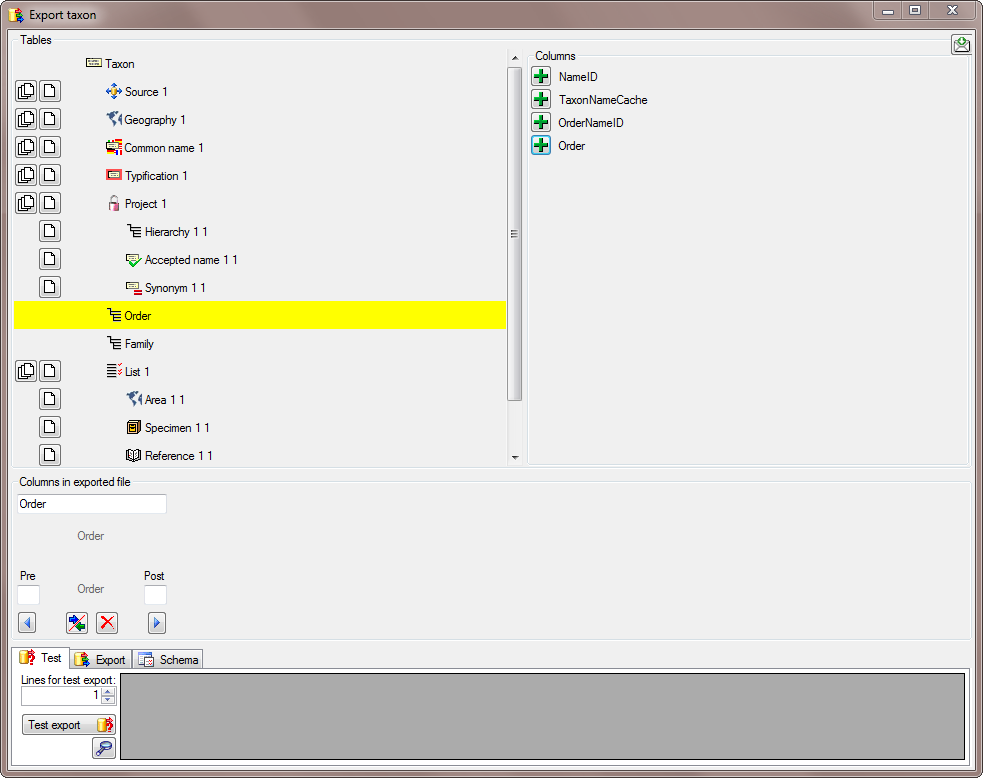
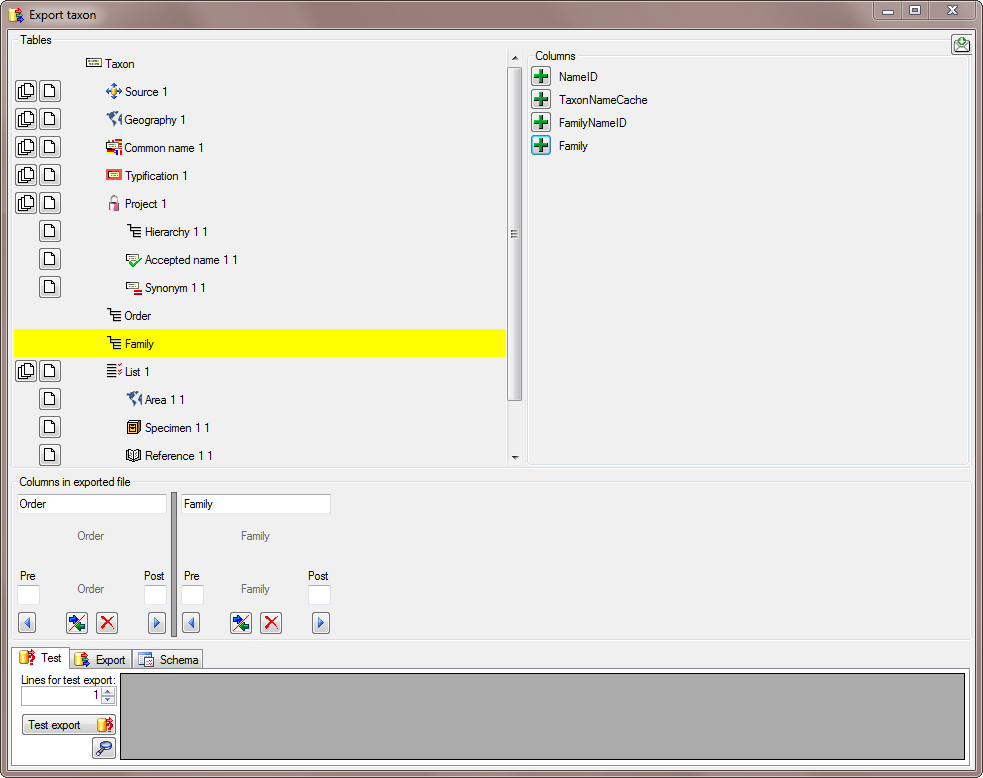
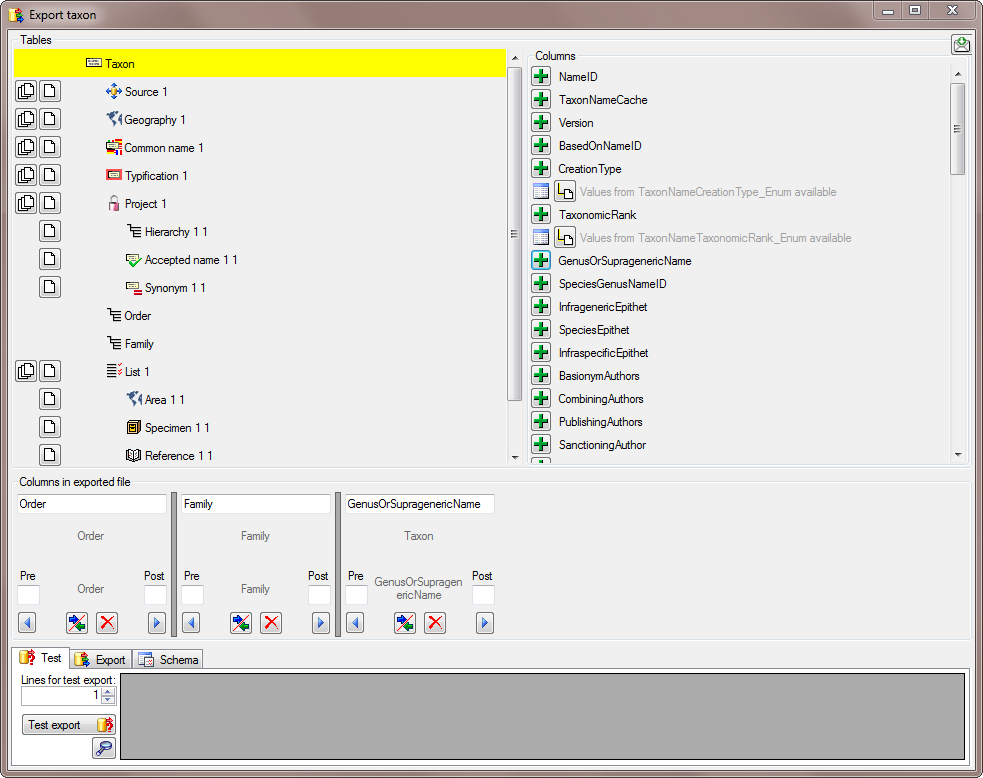
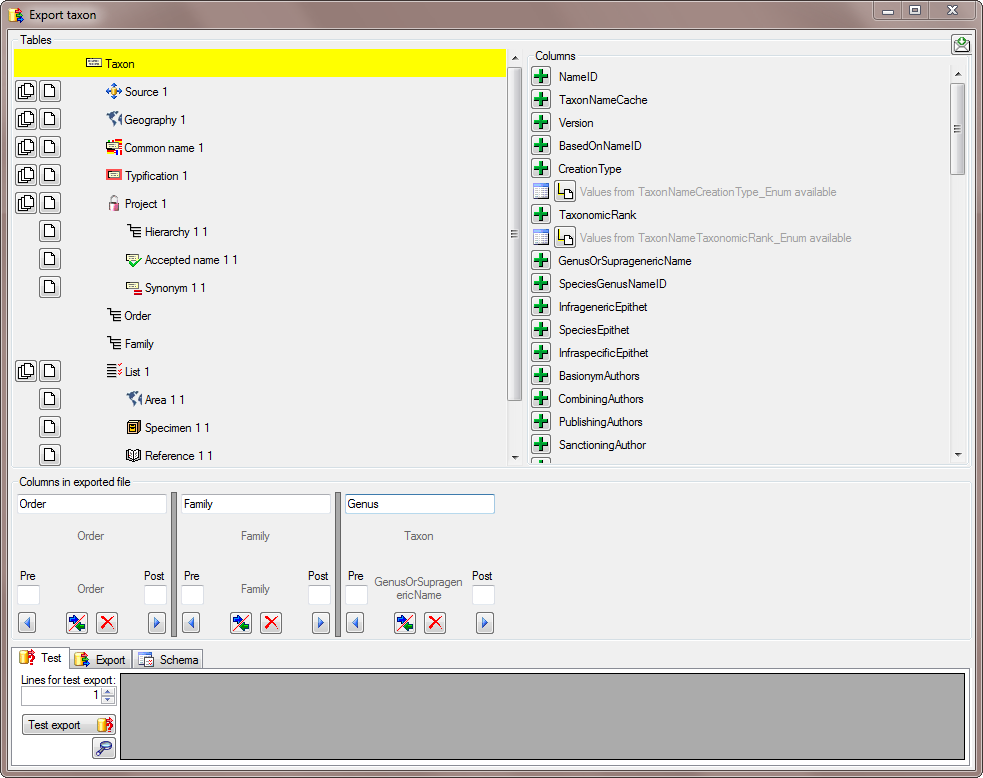
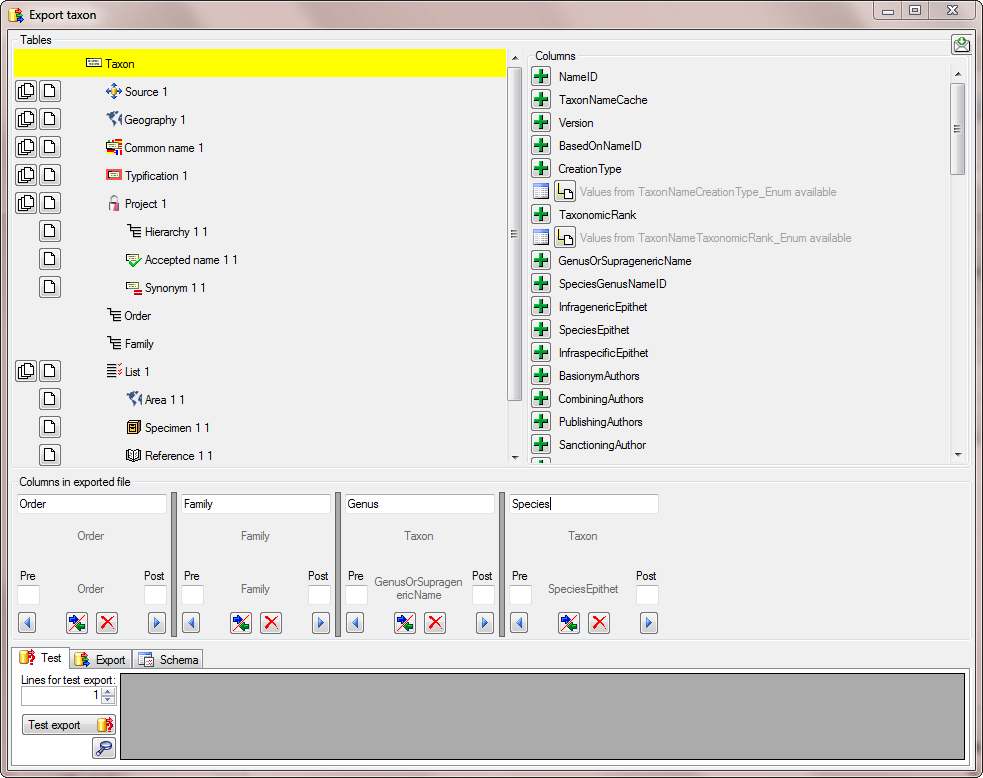
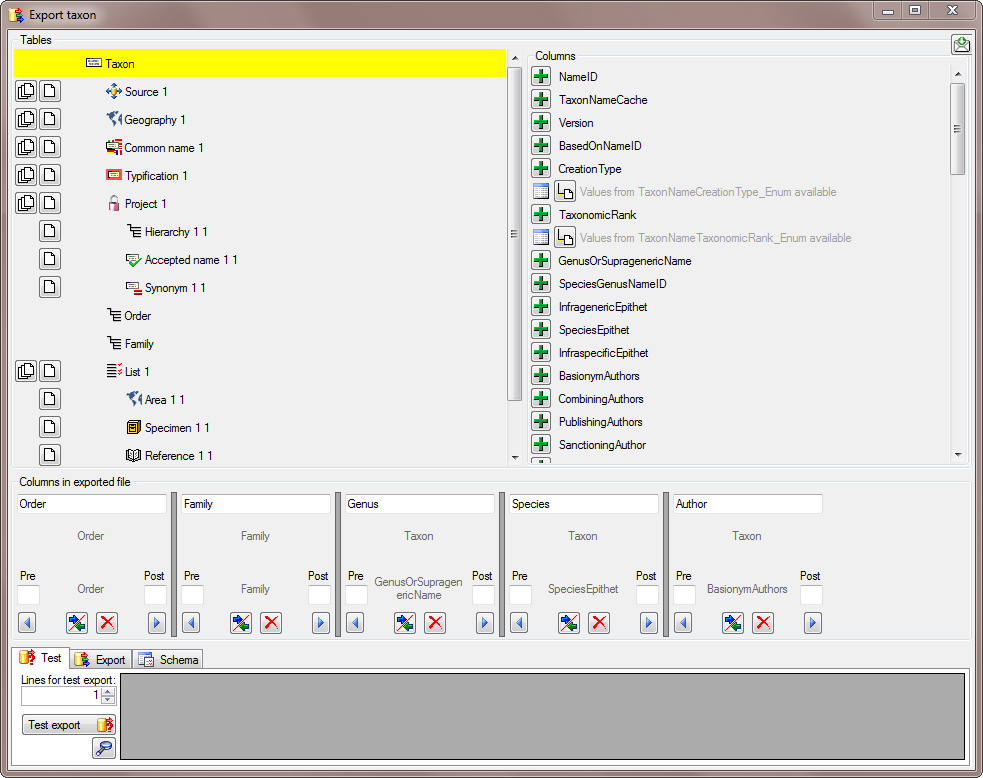
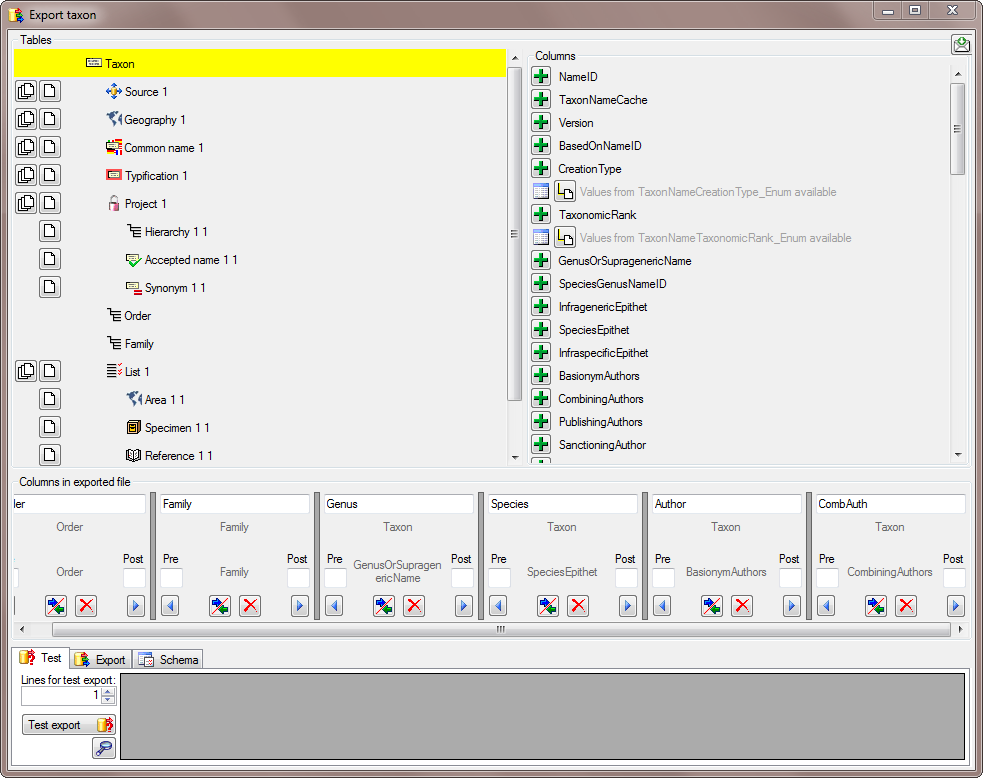
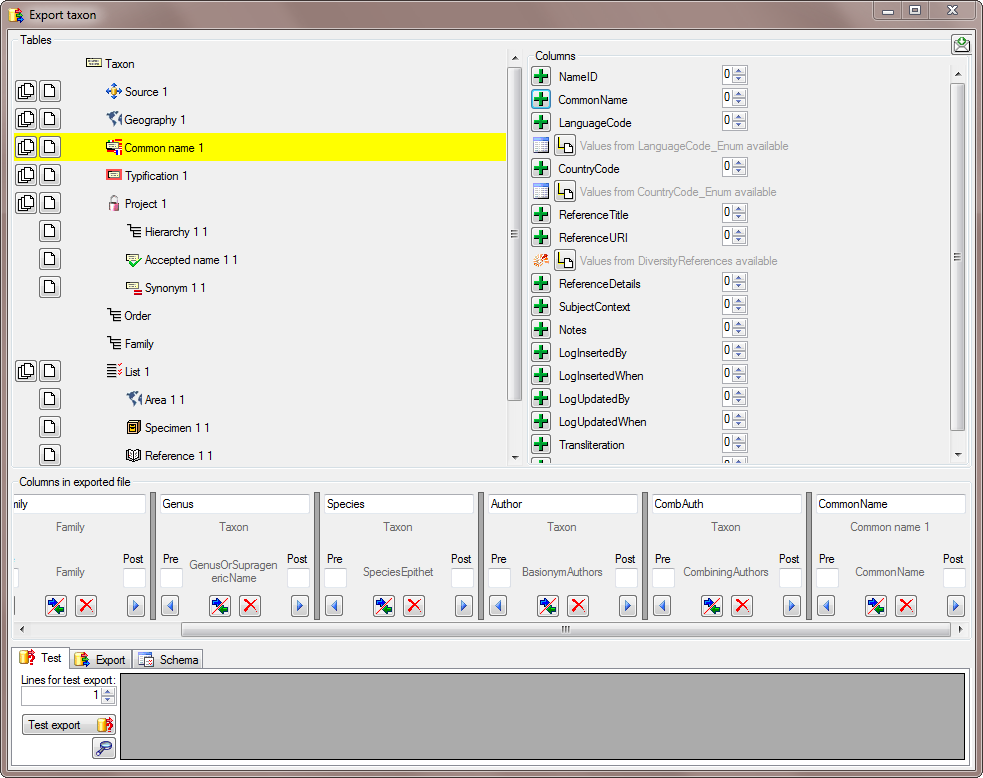
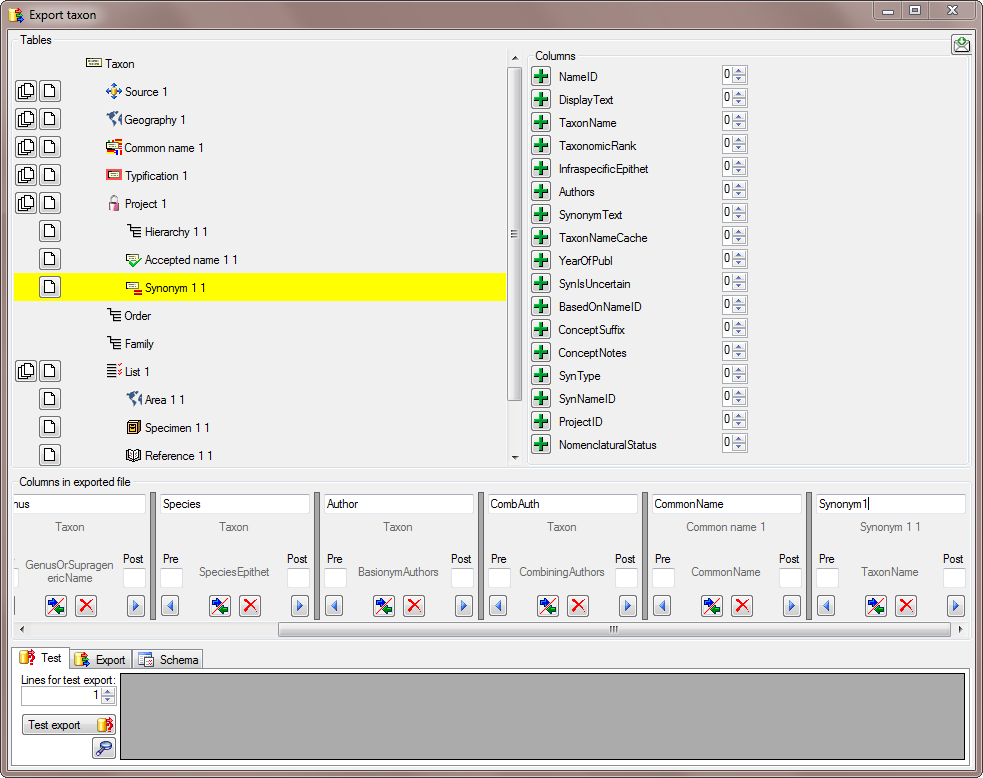
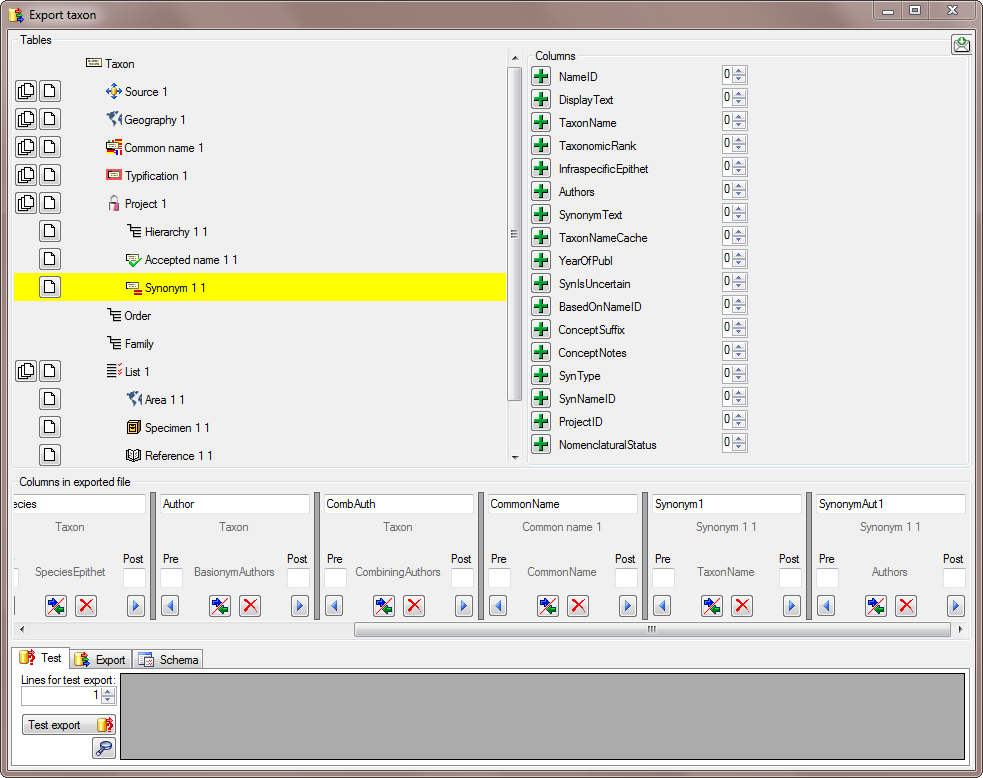
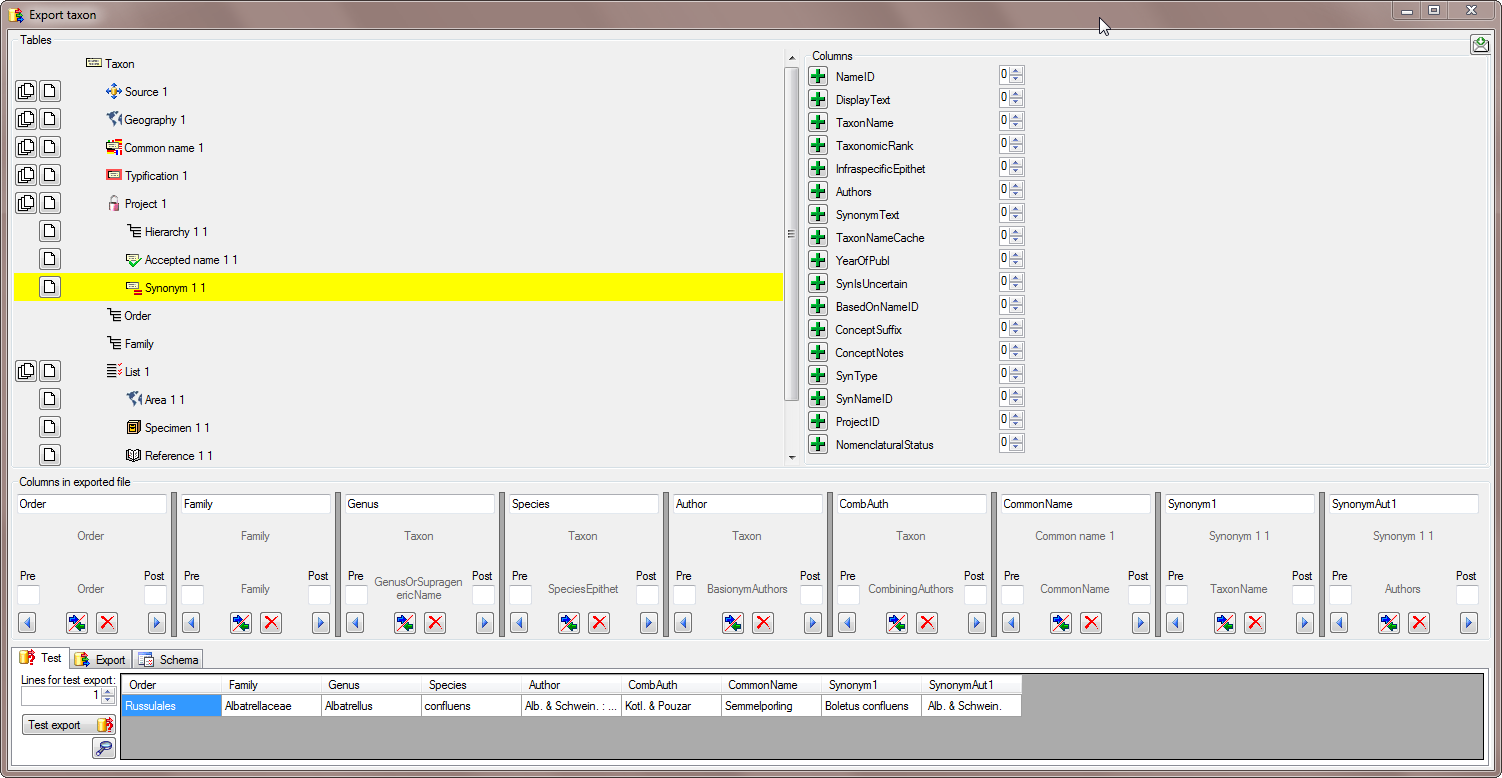
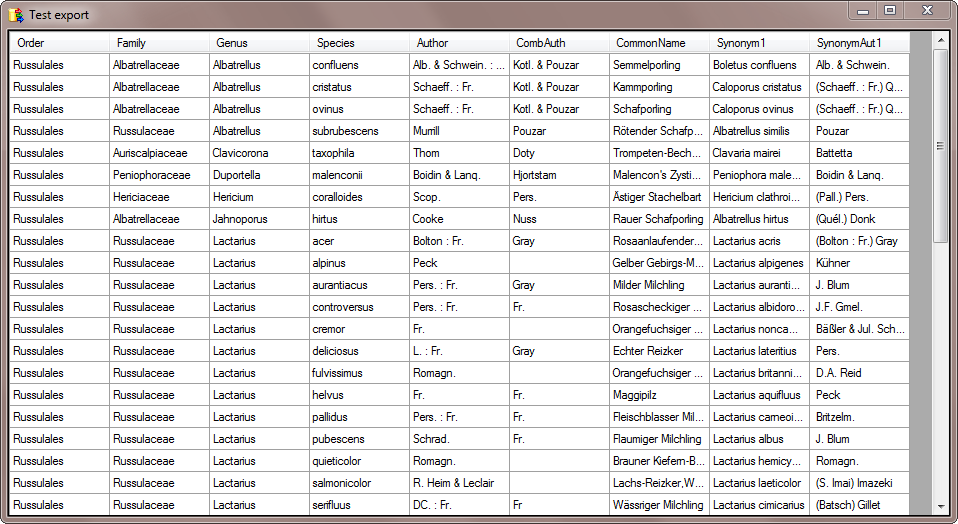
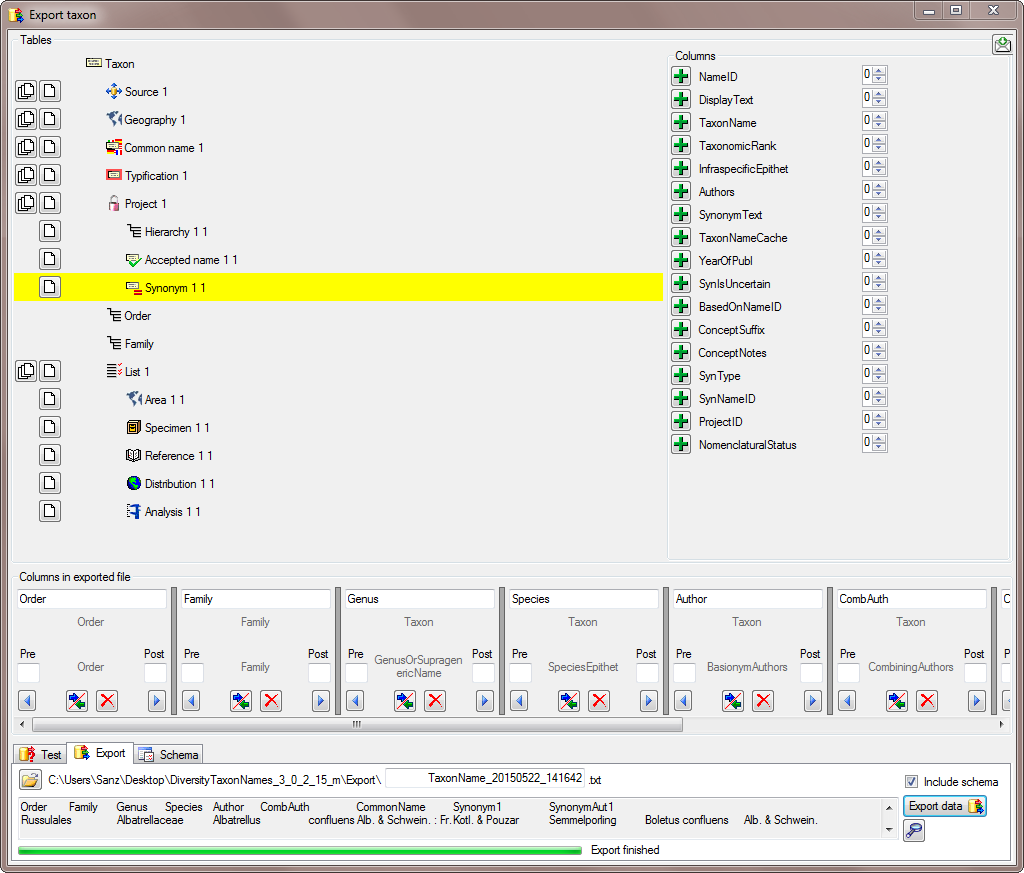

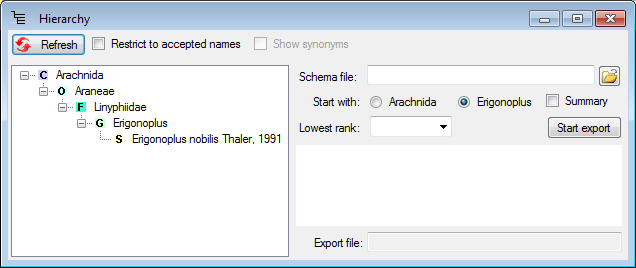
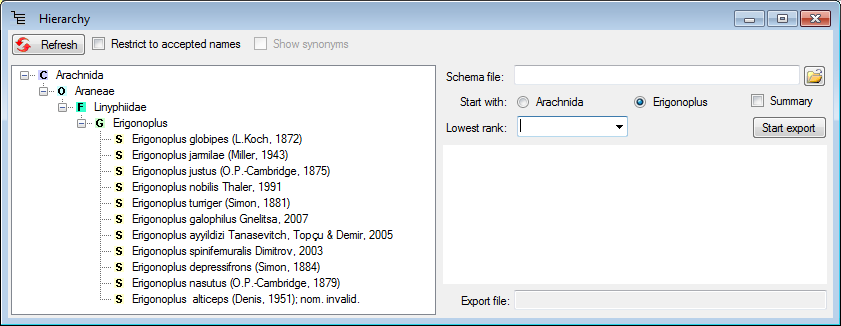
 button (see
below).
button (see
below).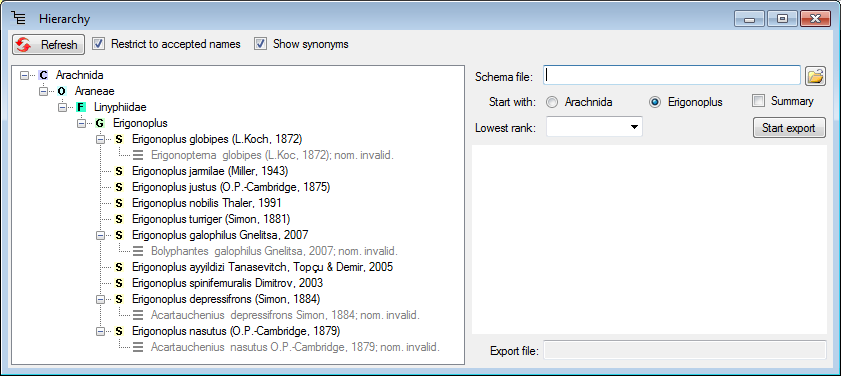
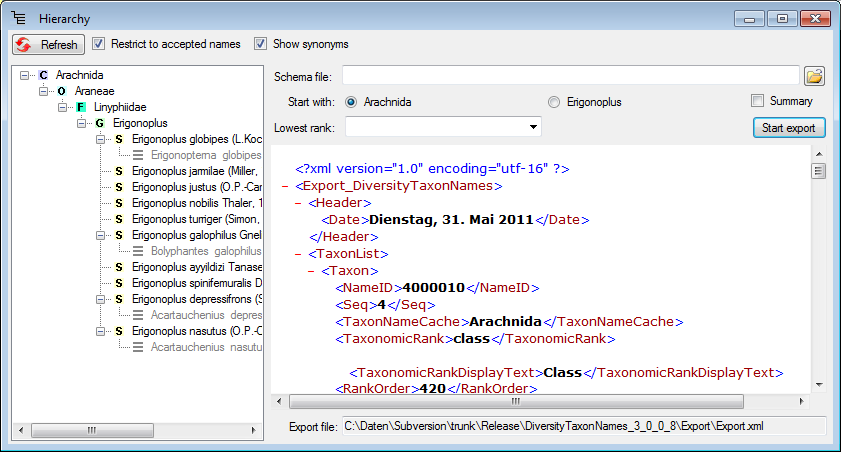
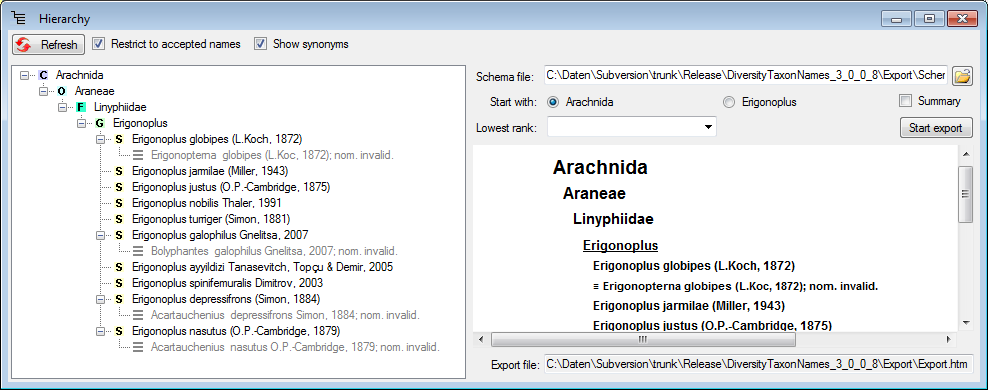
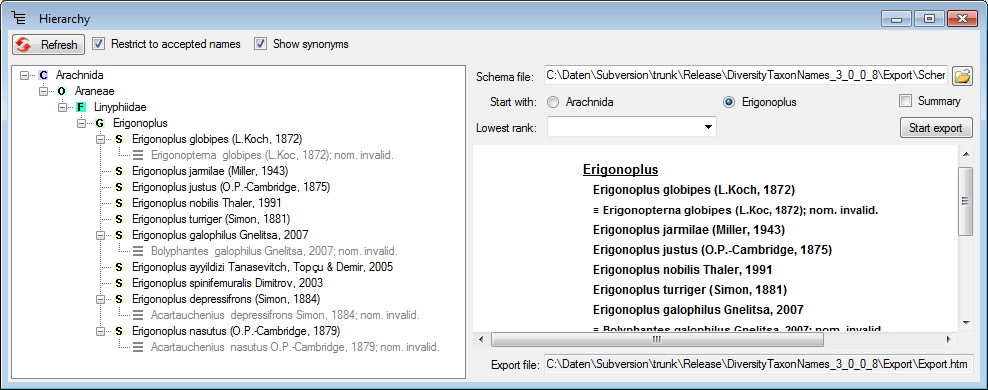
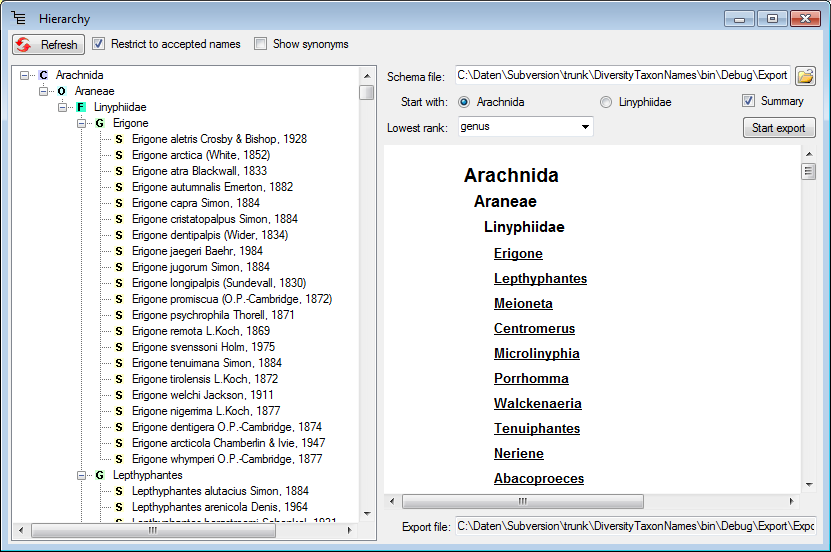
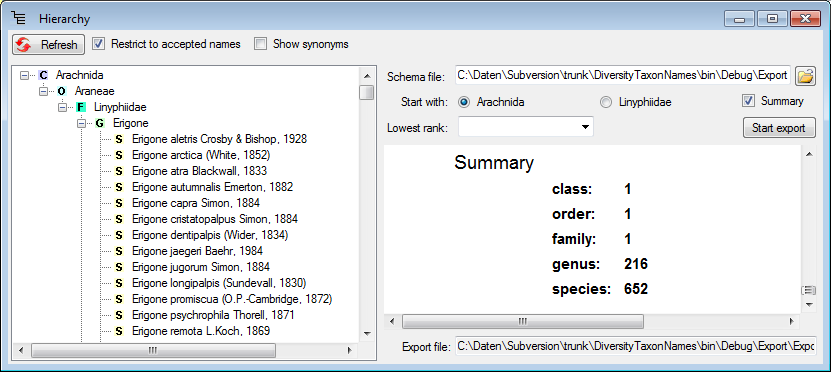
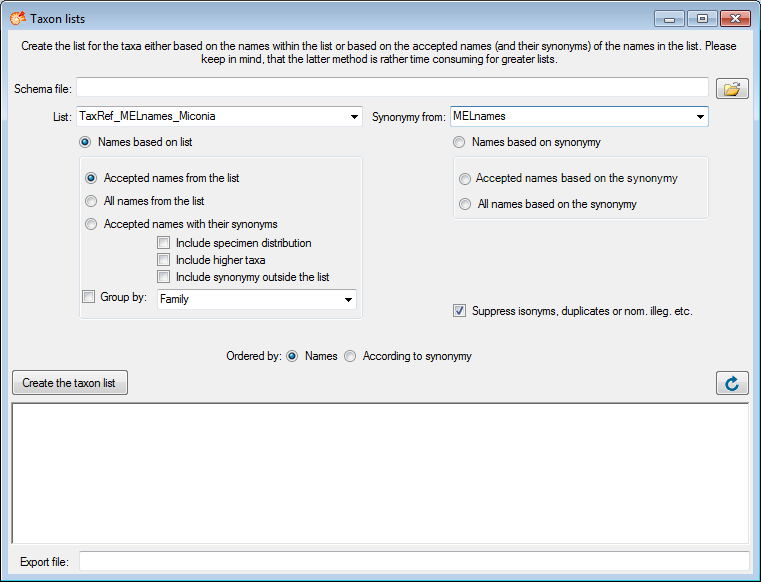
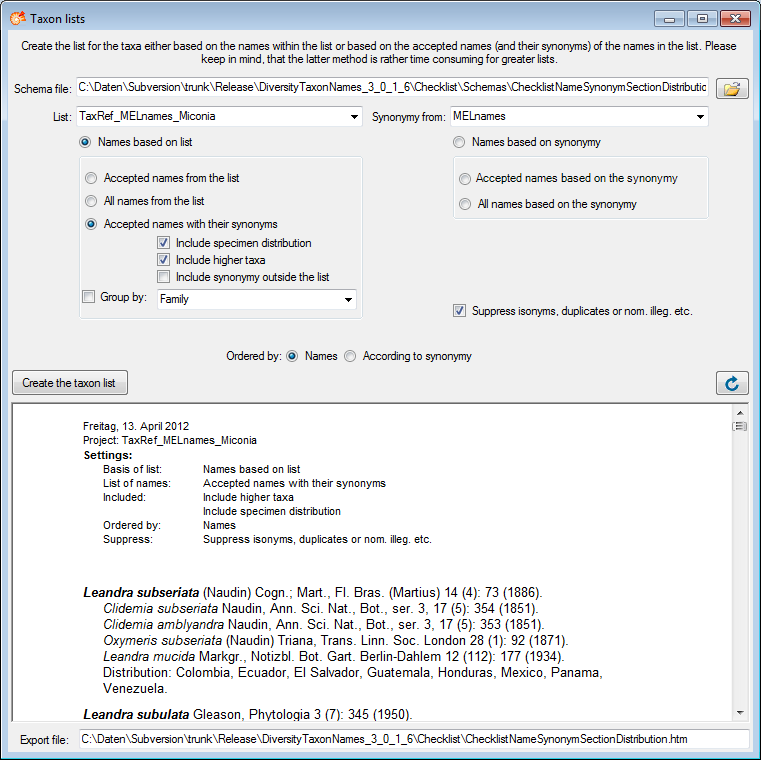
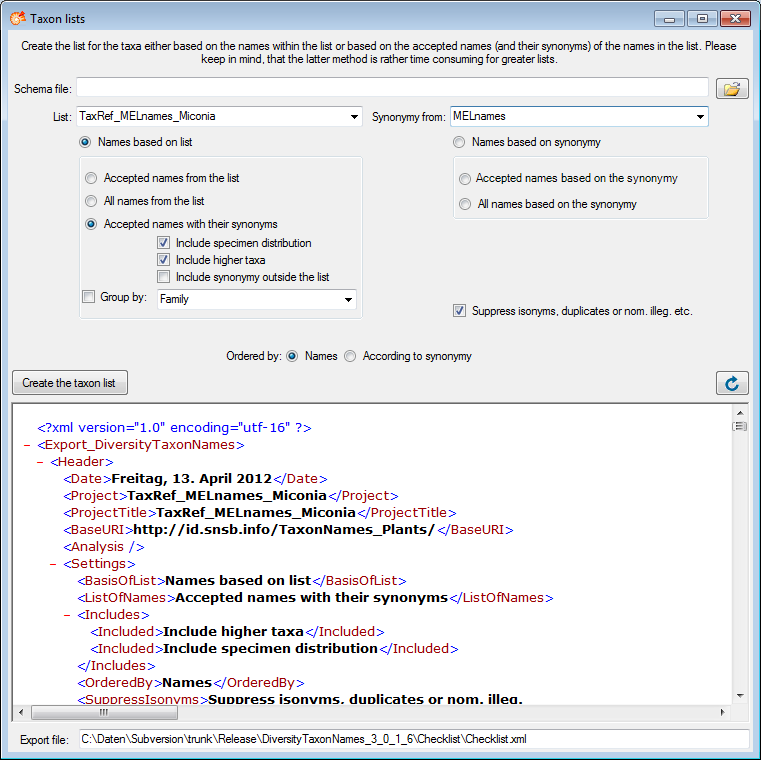
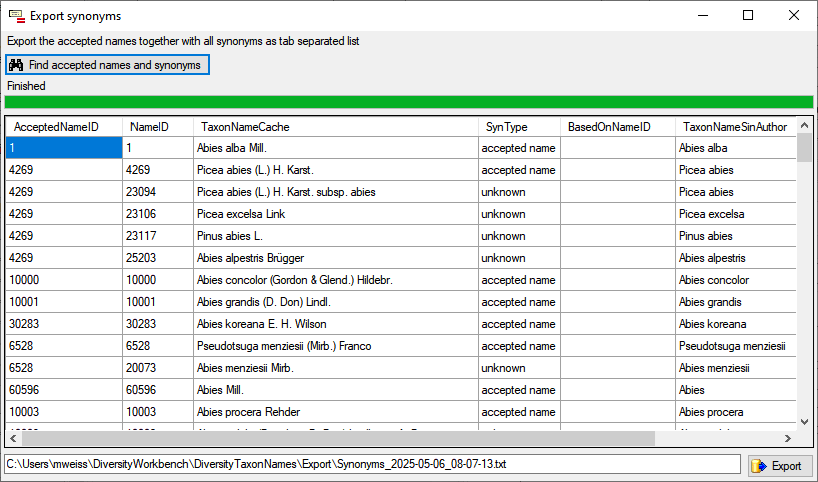



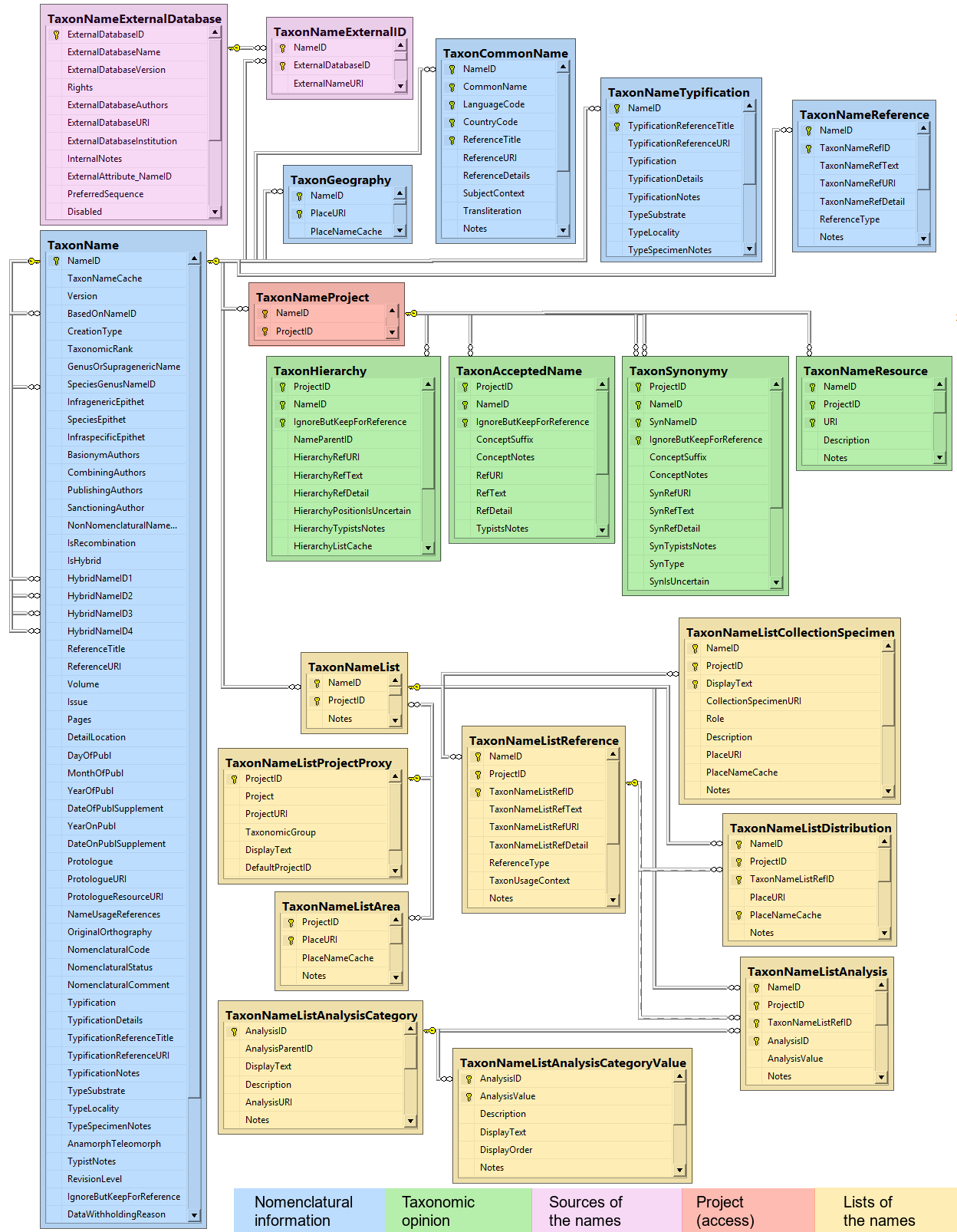



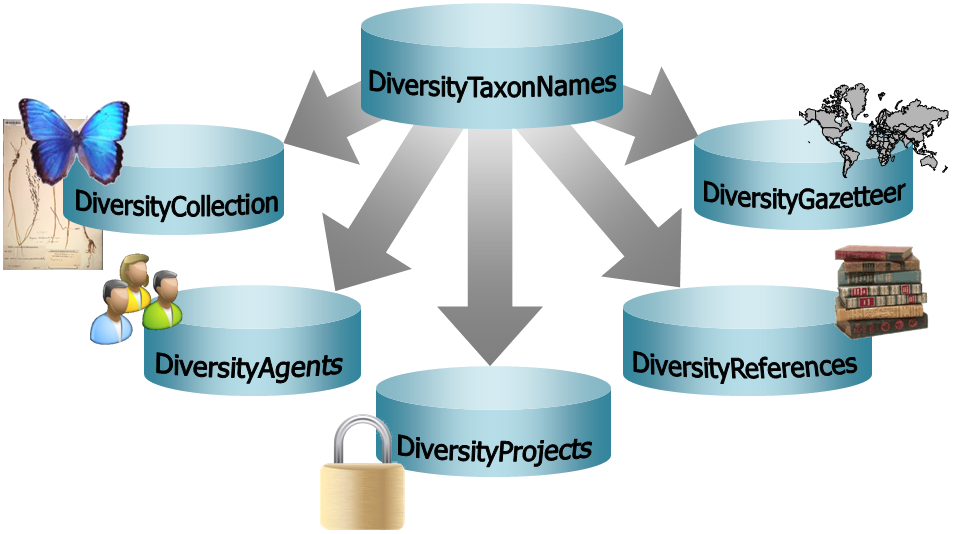
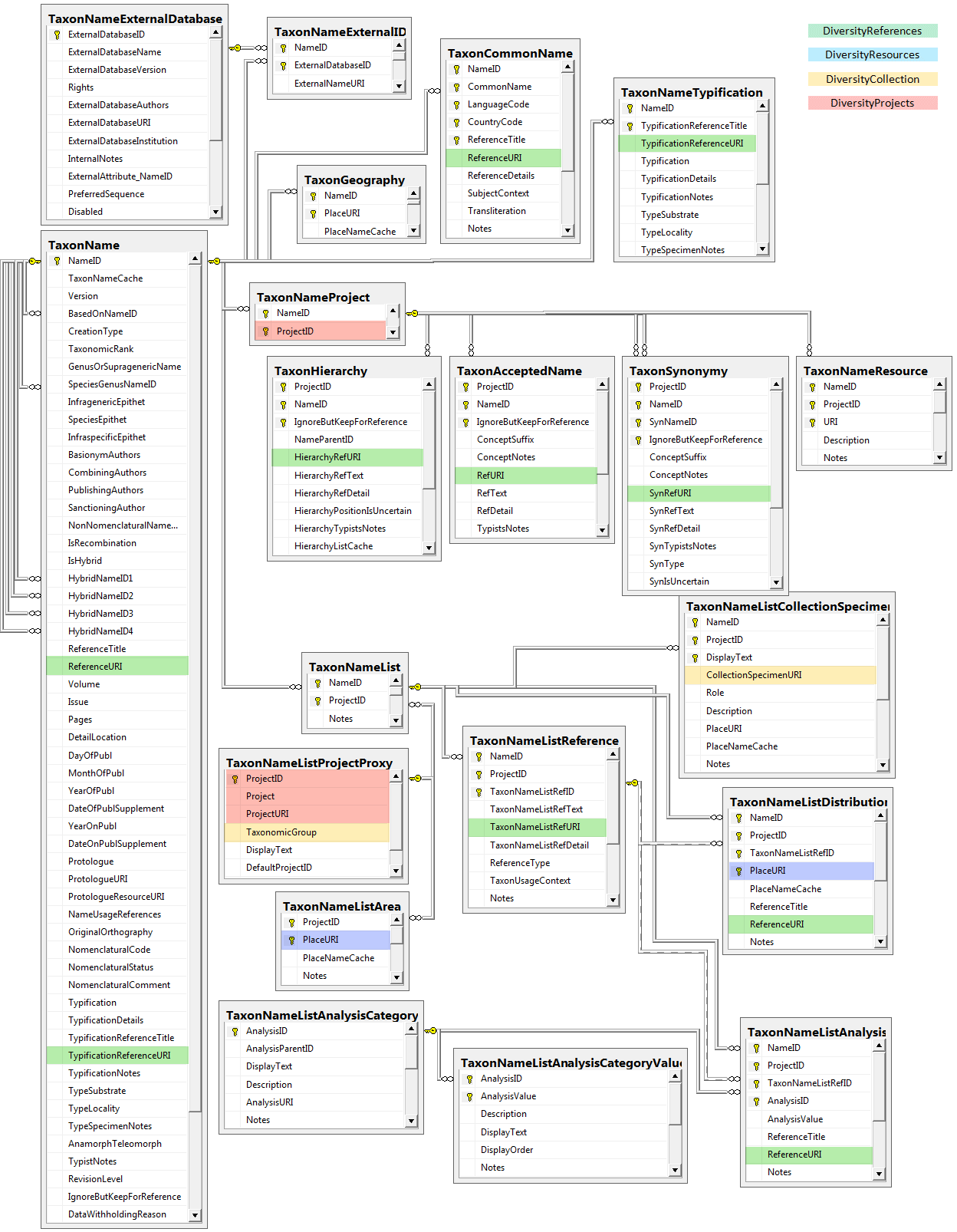

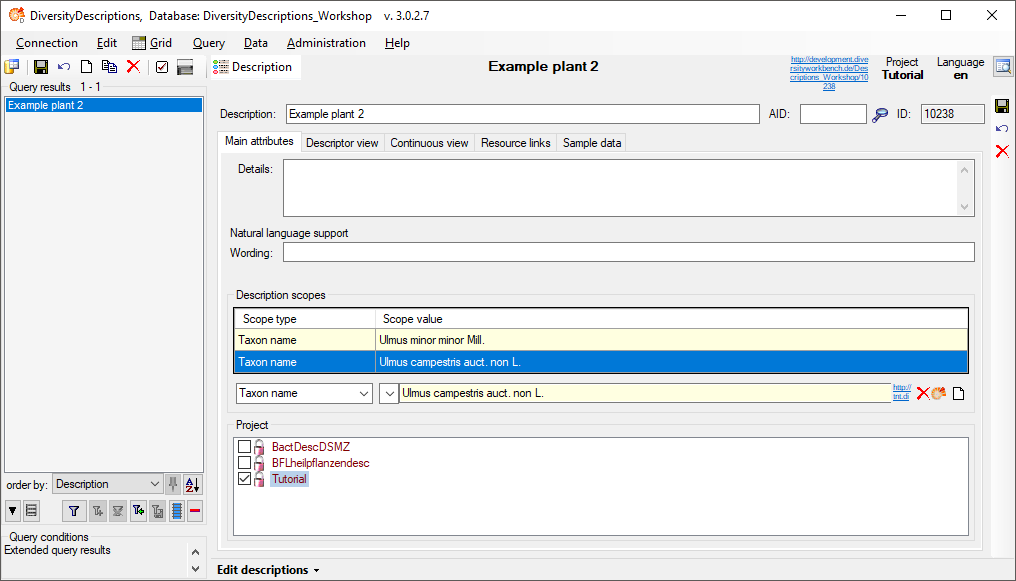

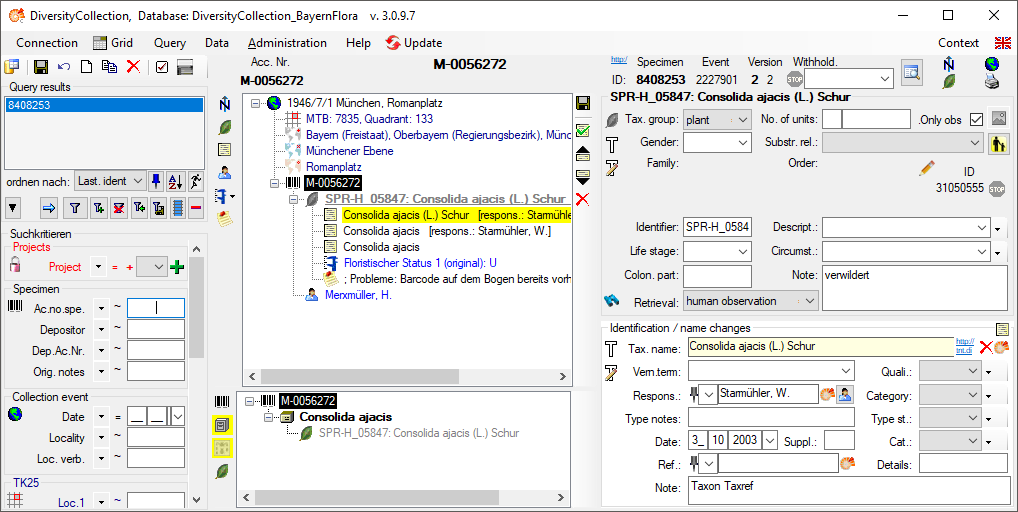
 button will be marked with a pink
background
button will be marked with a pink
background
