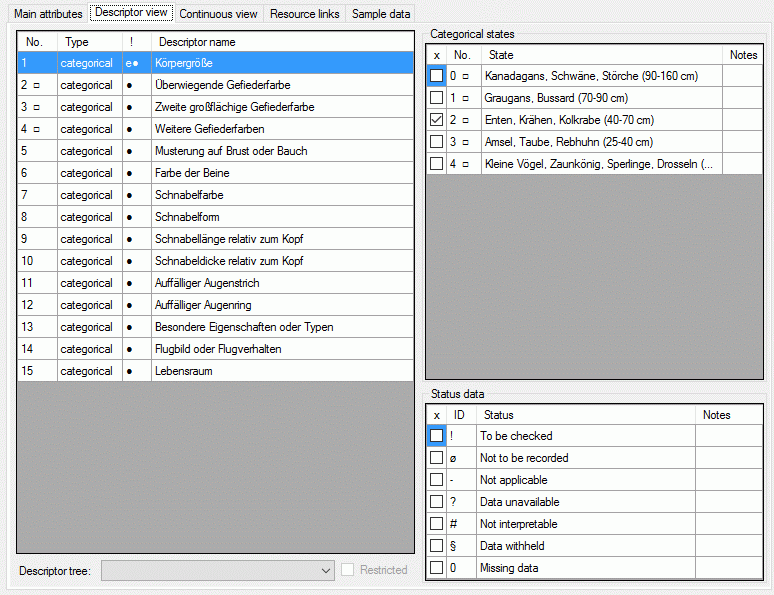

The left part of the descriptor view tab shows all descriptors available for the description's project. The entries in column "!" indicate some additional information about the descriptor:
If resources are linked to a descriptor, the entry is marked by a square (□) behind its sequency number and a tool tip appears when the mouse cursor is moved over the "No." column (see below). By double-clicking the sequence number a "Media view" window can be opened to view the resources.
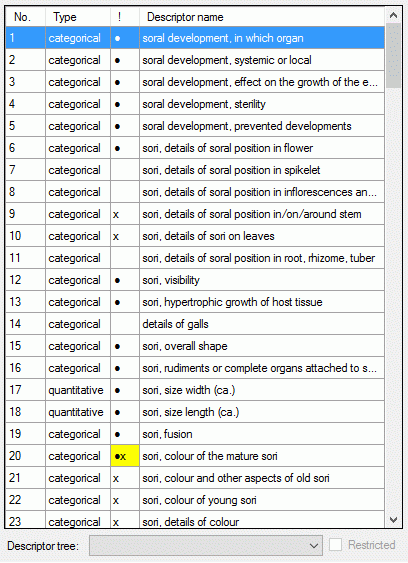
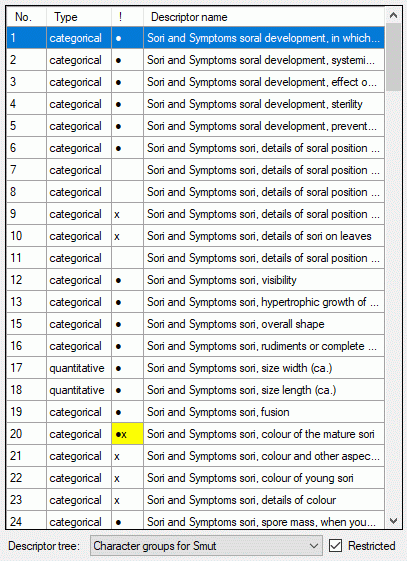
If for the project descriptor trees are defined, the Descriptor tree: drop-down list and Restricted check box are shown below the descriptor list (see image below left). If a descriptor tree is selected, the names of the descriptor tree node hierarchy is included in the descriptor names (see image below right). This feature is useful, if several descriptors with ambigious names exist the can now be distinguished. E.g. two descriptors "colour", one at descriptor tree node "Leaf", the other at descriptor tree node "Blossom" will become "Leaf colour" and "Blossom colour". If the descriptor name already includes the hierarchy, the duplicate parts will be filtered. In the mentioned example the descriptor names "Leaf colour" and "Blossom colour" will not be changed, if the descriptor tree is selected.
If additionally the Restricted option is set, only the descriptors included in the selected descriptor tree are displayed (see image below right, missing descriptor number 14). So you have the option to create several descriptor trees that include only special aspects of the description's property (e.g. morphology, molecular biology and so on) to get a clearly arranged descriptor list.
The right part of the descriptor view tab shows at the bottom the Status data values of the selected descriptor. It is possible to select several values for each descriptor. In the "Notes" column additional information concerning the selected data status may be entered. By a double click on the "Notes" entry a separate edit window can be opened.
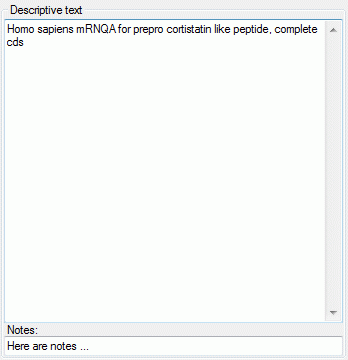
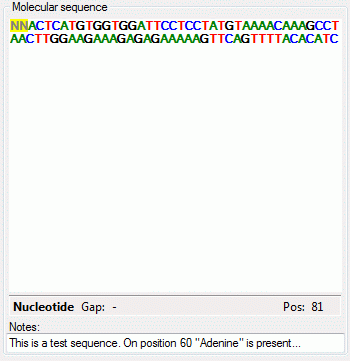
Depending on the descriptor type you can find at the top either Categorical states (see large picture above), Statistical measures, Descriptive text or Molecular sequence (see pictures below). In the "Notes" columns additional information concerning the specific categorical state rsp. statistical measure may be entered. By a double click on the "Notes" entry a separate edit window can be opened.
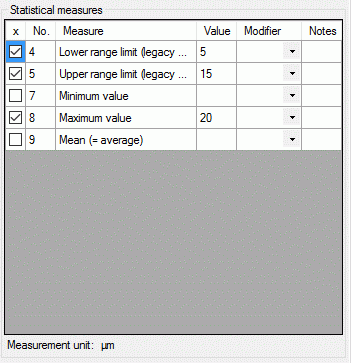
To enter a statistical measure value click in the "Value" field and enter the value, the check mark in field "x" will be set automatically. If recommended modifier values are defined (see "Editing the Descriptor" and "Editing the Project"), a value may be selected from the drop down list "Modifier". If no modifier values are available, the appropriate table column may be hidden. In the "Notes" field an additional text may be added. By double-clicking the "Notes" field the Extended text editor can be opened.
The descriptive text may be edited diretly in the window, which shows the pure text without any formatting. In the notes field an additional text may be entered. By double-clicking the text fields the Extended text editor can be opened.
The molecular sequence window displays in the status line below the edit area the sequence type ("Nucleotide" or "Protein"), the symbol length ("Len:") in case of proteine sequences, the gap symbol ("Gap:") if defined, the long text of the actual element and the position ("Pos:") (see image below). In case of three-letter proteine symbols the position is the number of the amino acid symbol, not the character position. By pressing the TAB key the cursor jumps to the next symbol.

When characters are entered in the edit area that are recognized as valid symbols, they will automatically be formatted. One-letter symbols are converted to upper case characters, three-letter symbols are converted according the "Xxx" schema, e.g. "Ala" for "Alanine". The sybols for the nucleotide bases "Adenine", "Cytosine", "Guanine", "Thymine" and "Uracil" are dispayed with different colors. Ambiguity symbols and gap symbols are shown in grey color. If the ambiguity symbols have not been enabled in the descriptor data, they will be displayed with yellow background. Symbols that have not been recognized are displayed with red background.
The molecular sequence window offers some additional functions
available in the sequence menu. It may
be opened by moving the mouse cursor over the control header ("Molecular sequence",
see window below left) or from the context menu (right-click on the sequence edit
area, see windows below right). With menu item
 Import you may read the seqeunce
data from a file and insert them to the description. With menu item
Import you may read the seqeunce
data from a file and insert them to the description. With menu item
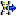 Export you may export the actual
sequence data to a file.
Export you may export the actual
sequence data to a file.
By double-clicking the molecular sequence area or from
sequence menu
 Open ->
Open ->
 Sequence a separate edit window may be opened (see image below). On
the top of the window there is the drop-down menu Insert symbol:
where a value may be selected from a list. After selecting the value press the [ENTER]
key or click on
Sequence a separate edit window may be opened (see image below). On
the top of the window there is the drop-down menu Insert symbol:
where a value may be selected from a list. After selecting the value press the [ENTER]
key or click on
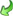 to insert the selected symbol at the cursor position.
to insert the selected symbol at the cursor position.
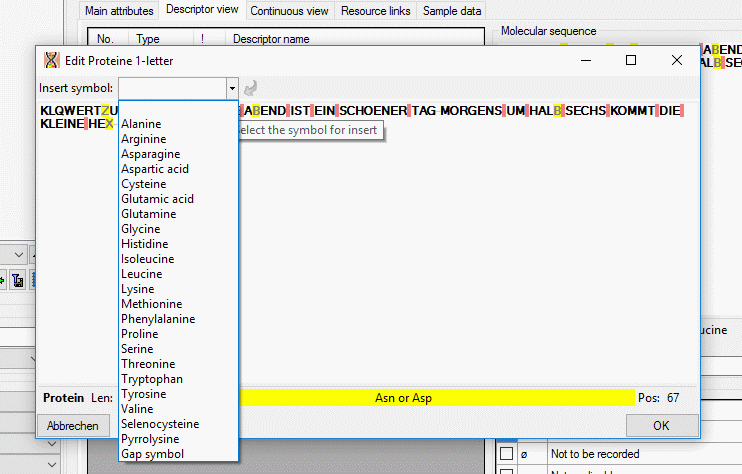
To select a categorical state set a check mark in field "x" with a mouse click. If recommended modifier or frequency values are defined (see Editing the Descriptor and Editing the Project), values may be selected from the drop down lists "Modifier" rsp. "Frequency". If no modifier or frequency values are available, the appropriate table column may be hidden. In the "Notes" field an additional text may be added. By double-clicking the "Notes" field the Extended text editor can be opened.
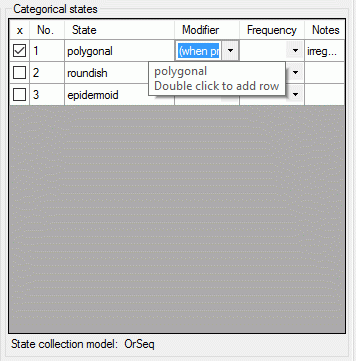
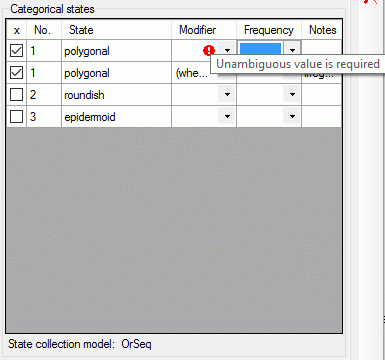
If a "Modifier" or "Frequency" value is selected, the multiple specification of a dedicated categorical state is possible. A tool tip offers to add a row by doouble-click, if mouse cursor is moved over the "State" field (left picture below, picture in the midddle after double click). Multiple specifications of one categorical state are only allowed if they have been assigned different modifier and frequency values. Inputs offending these restrictions will be ignored an a cell error will indicate the problem (right picture below).
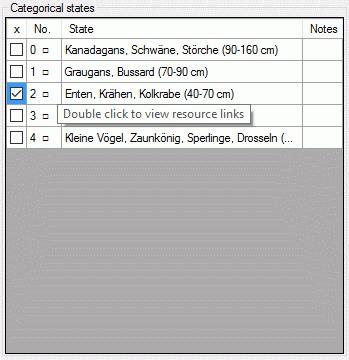
If resources are linked to a categorical state a tool tip appears when the mouse cursor is moved over the ID (left picture below). By double-clicking the ID a "Media view" window can be opened to view the resources (right picture below).
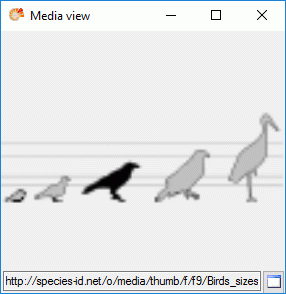
If more than one resources have been assigned to one entry, a slider will be visible at the bottom of the "Media view" window. To switch through the available resources the slider can be moved using the mouse or the right and left arrow keys (see below).
Continue with: