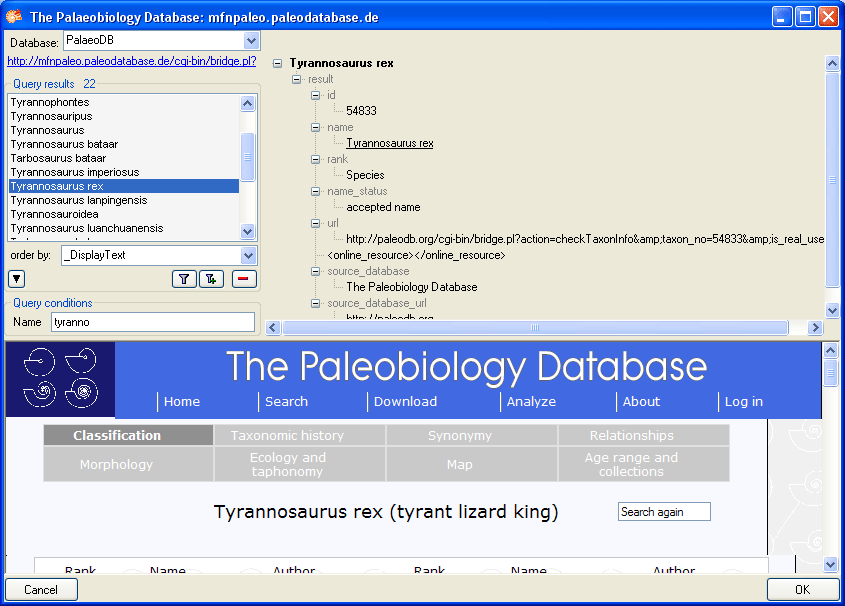
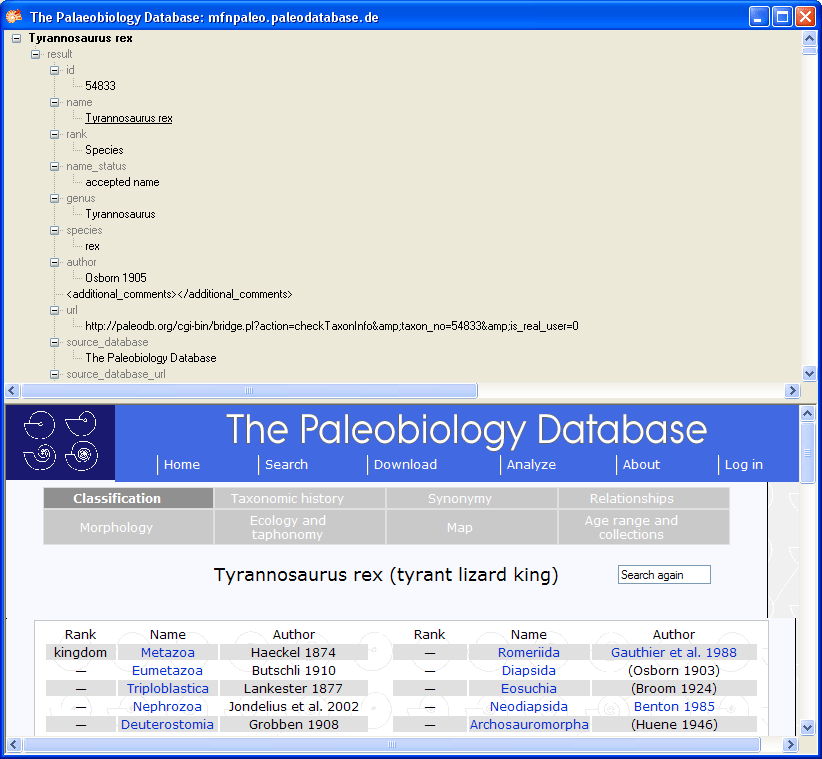
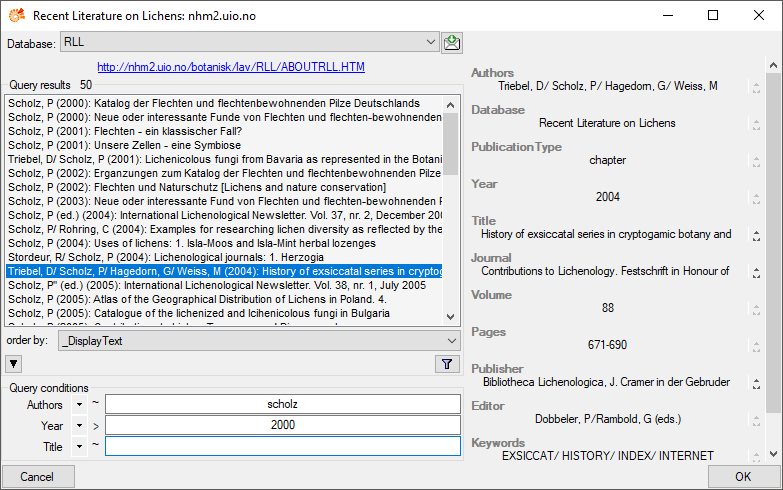
Subsections of Structure
Diversity Collection
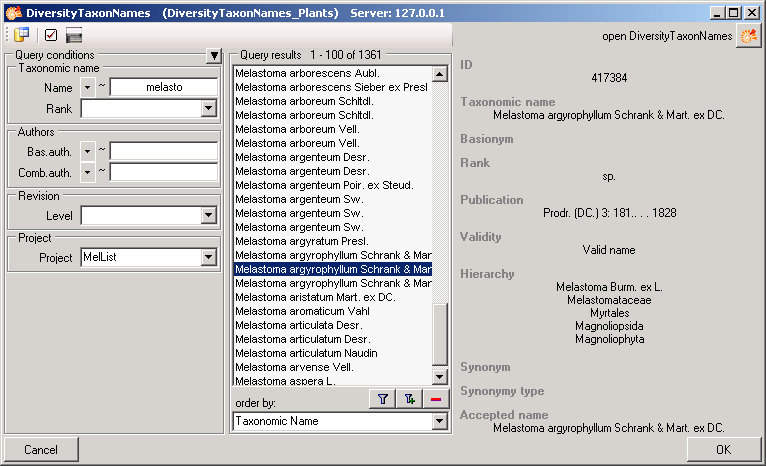
TABLES
The following objects are not included:
- Logging tables
- Enumeration tables
- System objects
- Objects marked as obsolete
- Previous versions of objects
Table Analysis
Analysis types used within the database
| Column |
Data type |
Description |
Nullable |
Relation |
| AnalysisID |
int |
ID of the analysis (primary key) |
NO |
- |
| AnalysisParentID |
int |
Analysis ID of the parent analysis, if it belongs to a certain type documented in this table |
YES |
Refers to table Analysis |
| DisplayText |
nvarchar (50) |
Name of the analysis as e.g. shown in user interface |
YES |
- |
| Description |
nvarchar (MAX) |
Description of the analysis |
YES |
- |
| MeasurementUnit |
nvarchar (50) |
The measurement unit used for the analysis, e.g. mm, µmol, kg |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on this analysis |
YES |
- |
| AnalysisURI |
varchar (255) |
URI referring to an external documentation of the analysis |
YES |
- |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entriesDefault value: (0) |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Table AnalysisResult
Value lists for analysis types with predefined values, e.g. “0, 1, 2, 3, …” for Red list category. Includes description etc. for the values in the list.
| Column |
Data type |
Description |
Nullable |
Relation |
| AnalysisID |
int |
ID of the analysis (primary key) |
NO |
Refers to table Analysis |
| AnalysisResult |
nvarchar (255) |
The categorized value of the analysis |
NO |
- |
| Description |
nvarchar (500) |
Description of enumerated object displayed in the user interface |
YES |
- |
| DisplayText |
nvarchar (50) |
Short abbreviated description of the object displayed in the user interface |
YES |
- |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
YES |
- |
| Notes |
nvarchar (500) |
Internal development notes on usage, definition, etc. of an enumerated object |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user to first enter (typ or import) the data.Default value: suser_sname() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Point in time when the data was first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data last.Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Point in time when this data was updated last.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table AnalysisTaxonomicGroup
The types of analysis which are available for a taxonomic group
| Column |
Data type |
Description |
Nullable |
Relation |
| AnalysisID |
int |
Analysis ID, foreign key of table Analysis. |
NO |
Refers to table Analysis |
| TaxonomicGroup |
nvarchar (50) |
Taxonomic group the organism, identified by this unit, belongs to. Groups listed in table CollTaxonomicGroup_Enum (= foreign key) |
NO |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table Annotation
Annotations to datasets in the database
| Column |
Data type |
Description |
Nullable |
Relation |
| AnnotationID |
int |
ID of the annotation (primary key) |
NO |
- |
| ReferencedAnnotationID |
int |
If an annotation refers to another annotation, the ID of the referred annotation |
YES |
Refers to table Annotation |
| AnnotationType |
nvarchar (50) |
The type of the annotation as defined in AnnotationType_Enum, e.g. ReferenceDefault value: N’Annotation' |
NO |
Refers to table AnnotationType_Enum |
| Title |
nvarchar (50) |
Title of the annotation |
YES |
- |
| Annotation |
nvarchar (MAX) |
The annotation entered by the user |
NO |
- |
| URI |
varchar (255) |
The complete URI address of a resource related to the annotation. May be link to a module, e.g. for the annotation type reference |
YES |
- |
| ReferenceDisplayText |
nvarchar (500) |
The title of the reference. If the entry is linked to an external module like DiversityReferences, the cached display text of the referenced data set |
YES |
- |
| ReferenceURI |
varchar (255) |
If the entry is linked to an external module like DiversityReferences, the link to the referenced data set |
YES |
- |
| SourceDisplayText |
nvarchar (500) |
The name of the source. If the entry is linked to an external module like DiversityAgents, the cached display text of the referenced data set |
YES |
- |
| SourceURI |
varchar (255) |
If the entry is linked to an external module like DiversityAgents, the link to the referenced data set |
YES |
- |
| IsInternal |
bit |
If an annotation is restricted to authorized users of the database |
YES |
- |
| ReferencedID |
int |
The ID of the data set in the table the annotation refers to |
NO |
- |
| ReferencedTable |
nvarchar (500) |
The name of the table the annotation refers to |
NO |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
trgInsAnnotation
updating the logging columns
Table AnonymCollector
Anonyms for collectors of whom the names should not be published
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectorsName |
nvarchar (400) |
The name of the collector, PK |
NO |
- |
| Anonymisation |
nvarchar (50) |
The anonymisation phrase for the collector |
YES |
- |
Table CacheDatabase2
Table holding the cache databases connected to the database
| Column |
Data type |
Description |
Nullable |
Relation |
| Server |
varchar (50) |
The name or IP of the server where the cache database is located |
NO |
- |
| DatabaseName |
varchar (50) |
The name of the cache database |
NO |
- |
| Port |
smallint |
The port of the server where the cache database is located |
NO |
- |
| Version |
varchar (50) |
The version of the cache database |
YES |
- |
Table CacheDescription
Table for temperary storage of description of database objects derived e.g. from tables Entity, EntityRepresentation etc.
| Column |
Data type |
Description |
Nullable |
Relation |
| TableName |
varchar (50) |
The name of the table |
NO |
- |
| ColumnName |
varchar (50) |
The name of the table column |
NO |
- |
| LanguageCode |
varchar (50) |
The language code for the descriptionDefault value: ’en-US' |
NO |
- |
| Context |
nvarchar (50) |
A context e.g. as definded in table EntityContext_EnumDefault value: ‘General’ |
NO |
- |
| DisplayText |
nvarchar (50) |
The text for the table or column as shown e.g. in a user interface |
YES |
- |
| Abbreviation |
nvarchar (20) |
The abbreviation for the table or column as shown e.g. in a user interface |
YES |
- |
| Description |
nvarchar (MAX) |
The description for the table column |
YES |
- |
| ID |
int |
A unique ID |
NO |
- |
| Type |
varchar (20) |
Type of the entryDefault value: ‘COLUMN’ |
YES |
- |
| Schema |
varchar (100) |
Schema of the entryDefault value: ‘dbo’ |
YES |
- |
Table Collection
The collections where the specimen are stored
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
NO |
- |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
YES |
Refers to table Collection |
| CollectionName |
nvarchar (255) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
NO |
- |
| CollectionAcronym |
nvarchar (10) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
YES |
- |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
YES |
- |
| AdministrativeContactAgentURI |
varchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
YES |
- |
| Description |
nvarchar (MAX) |
A short description of the collection |
YES |
- |
| Location |
nvarchar (255) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
YES |
- |
| LocationPlan |
varchar (500) |
URI or file name including path of the floor plan of the collection |
YES |
- |
| LocationPlanWidth |
float |
Width of location plan in meter for calculation of size by provided geometry |
YES |
- |
| LocationGeometry |
geometry (MAX) |
Geometry of the collection within the floor plan |
YES |
- |
| LocationHeight |
float |
Height from ground level, e.g. for the position of sensors |
YES |
- |
| LocationParentID |
int |
If the hierarchy of the location does not match the logical hierarchy, the ID of the parent location |
YES |
Refers to table Collection |
| LocationPlanDate |
datetime |
The date when the plan for the collection has been created |
YES |
- |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
YES |
- |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
YES |
- |
| Type |
nvarchar (50) |
Type of the collection, e.g. cupboard, drawer etc. |
YES |
Refers to table CollCollectionType_Enum |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table CollectionAgent
The collector(s) of CollectionSpecimens
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to ID of CollectionEvent (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimen |
| CollectorsName |
nvarchar (255) |
Name of the Collector |
NO |
- |
| CollectorsAgentURI |
varchar (255) |
The URI of the Agent, e.g. as stored within the module DiversityAgents |
YES |
- |
| CollectorsSequence |
datetime2 |
The order of collectors in a team. Automatically set by the database systemDefault value: sysdatetime() |
YES |
- |
| CollectorsNumber |
nvarchar (50) |
Number assigned to a specimen or a batch of specimens by the collector during the collection event (= ‘field number’) |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the collector, e.g. if the name is uncertain |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
trgInsCollectionAgent
Setting the version of the dataset
Table CollectionEvent
The event where and when the specimen were collected
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionEventID |
int |
Unique ID for the table CollectionEvent (= primary key) |
NO |
- |
| Version |
int |
The version of the data set. Automatically set by the system.Default value: (1) |
NO |
- |
| SeriesID |
int |
The ID of the related expedition. Relates to the PK of the table CollectionExpedition (foreign key). |
YES |
Refers to table CollectionEventSeries |
| CollectorsEventNumber |
nvarchar (50) |
Number assigned to a collection event by the collector (= ‘field number’) |
YES |
- |
| CollectionDate |
datetime |
The cached date of the collection event calulated from the entries in CollectionDay, -Month and -Year. |
YES |
- |
| CollectionDay |
tinyint |
The day of the date of the event or when the collection event started |
YES |
- |
| CollectionMonth |
tinyint |
The month of the date of the event or when the collection event started |
YES |
- |
| CollectionYear |
smallint |
The year of the date of the event or when the collection event started |
YES |
- |
| CollectionEndDay |
tinyint |
The day of the date of the event or when the collection event ended |
YES |
- |
| CollectionEndMonth |
tinyint |
The month of the date of the event or when the collection event ended |
YES |
- |
| CollectionEndYear |
smallint |
The year of the date of the event or when the collection event ended |
YES |
- |
| CollectionDateSupplement |
nvarchar (100) |
Verbal or additional collection date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. The end date, if the collection event comprises a period. The time of the event, if necessary. |
YES |
- |
| CollectionDateCategory |
nvarchar (50) |
Category of the date of the identification e.g. “system”, “estimated” (= foreign key, see in table CollEventDateCategory_Enum) |
YES |
Refers to table CollEventDateCategory_Enum |
| CollectionTime |
varchar (50) |
The time of the event or when the collection event started |
YES |
- |
| CollectionTimeSpan |
varchar (50) |
The time span e.g. in seconds of the collection event |
YES |
- |
| LocalityDescription |
nvarchar (MAX) |
Locality description of the locality exactly as written on the original label (i.e. without corrections during data entry) |
YES |
- |
| LocalityVerbatim |
nvarchar (MAX) |
Locality as given in historical context, documents and labels |
YES |
- |
| HabitatDescription |
nvarchar (MAX) |
Geo-ecological description of the locality exactly as written on the original label (i.e. without corrections during data entry) |
YES |
- |
| ReferenceTitle |
nvarchar (255) |
The title of the publication where the collection event was published. Note that this is only a cached value where ReferenceURI is present |
YES |
- |
| ReferenceURI |
varchar (255) |
URI (e.g. LSID) of the source publication where the collection event is published, may e.g. refer to the module DiversityReferences |
YES |
- |
| ReferenceDetails |
nvarchar (50) |
The exact location within the reference, e.g. pages, plates |
YES |
- |
| CollectingMethod |
nvarchar (MAX) |
Description of the method used for collecting the samples, e.g. traps, moist chambers, drag net |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the collection event |
YES |
- |
| CountryCache |
nvarchar (50) |
The country where the collection event took place. Cached value derived from an entry in CollectionEventLocalisation |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| DataWithholdingReasonDate |
nvarchar (50) |
The reason for withholding the collection date |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionEventSeries
- CollEventDateCategory_Enum
trgInsCollectionEvent
Setting the date in case of valid date columns
Table CollectionEventImage
The images showing the collection site resp. place of the observations
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionEventID |
int |
Part of primay key, refers to unique ID for the table CollectionEvent (= foreign key) |
NO |
Refers to table CollectionEvent |
| URI |
varchar (255) |
The complete URI address of the image. This is only a cached value, if ResourceID is available and referring to the module DiversityResources |
NO |
- |
| ResourceURI |
varchar (255) |
The URI of the resource (e.g. see module DiversityResources) |
YES |
- |
| ImageType |
nvarchar (50) |
Type of the image, e.g. map |
YES |
Refers to table CollEventImageType_Enum |
| Notes |
nvarchar (MAX) |
Notes to this image concerning the CollectionEvent |
YES |
- |
| Description |
xml (MAX) |
Description of the image |
YES |
- |
| Title |
nvarchar (500) |
Title of the resource |
YES |
- |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
- |
| CreatorAgent |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
- |
| CreatorAgentURI |
varchar (255) |
Link to the module DiversityAgents |
YES |
- |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
- |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e. g. Creative Common Licenses |
YES |
- |
| InternalNotes |
nvarchar (500) |
Internal notes which should not be published e.g. on websites |
YES |
- |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
- |
| LicenseHolderAgentURI |
nvarchar (500) |
The link to a module containing futher information on the person or institution holding the license |
YES |
- |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionEvent
- CollEventImageType_Enum
trgInsCollectionEventImage
Setting the version of the dataset
Table CollectionEventLocalisation
The geographic localisation of a CollectionEvent
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionEventID |
int |
Refers to the ID of CollectionEvent (= foreign key and part of primary key) |
NO |
Refers to table CollectionEvent |
| LocalisationSystemID |
int |
Refers to the ID of LocalisationSystem (= foreign key and part of primary key) |
NO |
Refers to table LocalisationSystem |
| Location1 |
nvarchar (255) |
Either a named location selected from a thesaurus (e.g. ‘Germany, Bavaria, Kleindingharting’) or altitude range or other values (e. g. 100-200 m) |
YES |
- |
| Location2 |
nvarchar (255) |
Corresponding value to Location1 e.g. ID or URI of gazetteer or thesaurus |
YES |
- |
| LocationAccuracy |
nvarchar (50) |
The accuracy of the determination of this locality |
YES |
- |
| LocationNotes |
nvarchar (MAX) |
Notes on the location |
YES |
- |
| DeterminationDate |
smalldatetime |
Date of the determination of the geographical localisation |
YES |
- |
| DistanceToLocation |
varchar (50) |
Distance from the specified place to the real location of the collection site (m) |
YES |
- |
| DirectionToLocation |
varchar (50) |
Direction from the specified place to the real location of the collection site (Degrees rel. to north) |
YES |
- |
| ResponsibleName |
nvarchar (255) |
The name of the agent (person or organization) responsible for this entry. |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or organisation responsible for the data (see e.g. module DiversityAgents) |
YES |
- |
| Geography |
geography |
The geography of the localisation |
YES |
- |
| RecordingMethod |
nvarchar (500) |
The method or device used for the recording of the localisation |
YES |
- |
| AverageAltitudeCache |
float |
Calculated altitude as parsed from the location fields |
YES |
- |
| AverageLatitudeCache |
float |
Calculated latitude as parsed from the location fields |
YES |
- |
| AverageLongitudeCache |
float |
Calculated longitude as parsed from the location fields |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionEvent
- LocalisationSystem
trgInsCollectionEventLocalisation
Setting missing geographical values on base of given values
Table CollectionEventMethod
The methods used during a collection event
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionEventID |
int |
Refers to ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionEvent |
| MethodID |
int |
ID of the setting, part of primary key |
NO |
Refers to table Method |
| MethodMarker |
nvarchar (50) |
A marker for the method, part of primary keyDefault value: ‘1’ |
NO |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table CollectionEventParameterValue
The values of the parameter of the methods used within a collection event
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionEventID |
int |
Unique ID for the table CollectionEvent (= foreign key and part of primary key) |
NO |
Refers to table CollectionEventMethod |
| MethodID |
int |
ID of the method tool. Referes to table Method (= foreign key and part of primary key) |
NO |
Refers to table CollectionEventMethod and table Parameter |
| MethodMarker |
nvarchar (50) |
A marker for the method, part of primary keyDefault value: ‘1’ |
NO |
Refers to table CollectionEventMethod |
| ParameterID |
int |
ID of the parameter tool. Referes to table Parameter (= foreign key and part of primary key) |
NO |
Refers to table Parameter |
| Value |
nvarchar (MAX) |
The value of the parameter, if different of the default value as documented in the table Parameter |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes concerning the value of the parameter |
YES |
- |
| LogInsertedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
YES |
- |
Depending on:
- CollectionEventMethod
- Parameter
Table CollectionEventProperty
A property of a collection site, e.g. exposition, slope, vegetation. May refer to Diversity Workbench module DiversityScientificTerms
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionEventID |
int |
Refers to the ID of CollectionEvent (= foreign key and part of primary key) |
NO |
Refers to table CollectionEvent |
| PropertyID |
int |
The ID of the property of the collection site, foreign key, see table Property |
NO |
Refers to table Property |
| DisplayText |
nvarchar (255) |
The text for the property as shown e.g. in a user interface |
YES |
- |
| PropertyURI |
varchar (255) |
URI referring to an external data source e.g. DiversityTerminology |
YES |
- |
| PropertyHierarchyCache |
nvarchar (MAX) |
A cached text of the complete name of the descriptor including superior categories, if present |
YES |
- |
| PropertyValue |
nvarchar (255) |
The value of a captured feature, e.g. temperature, pH, vegetation etc. If there is a range, this is the lower or first value |
YES |
- |
| ResponsibleName |
nvarchar (255) |
The name of the agent (person or organization) responsible for this entry. |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or organisation responsible for the data (see e.g. module DiversityAgents) |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the property of the colletion site. |
YES |
- |
| AverageValueCache |
float |
For numeric values - a cached average value according to the |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
trgInsCollectionEventProperty
Setting the version of the dataset
Table CollectionEventRegulation
Regulation applied to a collection event
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionEventID |
int |
Part of primay key, refers to unique ID for the table CollectionEvent (= foreign key) |
NO |
Refers to table CollectionEvent |
| Regulation |
nvarchar (400) |
Regulation as defined in the table Regulation. Used to ensure, that user checked correct entry with authorized stuff |
NO |
- |
| TransactionID |
int |
Refers to unique TransactionID for the table Transaction (= foreign key) |
YES |
Refers to table Transaction |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionEvent
- Transaction
Table CollectionEventSeries
The series whithin which collection events take place
| Column |
Data type |
Description |
Nullable |
Relation |
| SeriesID |
int |
Primary key. The ID for this series of collection events (= primary key) |
NO |
- |
| SeriesParentID |
int |
The ID of the superior series of collection events |
YES |
Refers to table CollectionEventSeries |
| Description |
nvarchar (MAX) |
The description of the series of collection events as it will be printed on e.g. the label |
NO |
- |
| SeriesCode |
nvarchar (50) |
The user defined code for a series of collection events |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on this series of collection events |
YES |
- |
| Geography |
geography |
The geography of the series of collection events |
YES |
- |
| DateStart |
datetime |
Point in time when the series of collection events started |
YES |
- |
| DateEnd |
datetime |
Point in time when the series of collection events ended |
YES |
- |
| DateCache |
datetime |
The first date of the depending events, used for sorting the expeditions [controlled by the database] |
YES |
- |
| DateSupplement |
nvarchar (100) |
Verbal or additional collection date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. The end date, if the collection event series comprises a period. The time of the event, if necessary. |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Table CollectionEventSeriesDescriptor
The Descriptors for the CollectionEventSeries
| Column |
Data type |
Description |
Nullable |
Relation |
| SeriesID |
int |
Unique ID for the CollectionEventSeries (foreign key + part of primary key) |
NO |
Refers to table CollectionEventSeries |
| DescriptorID |
int |
Unique ID for the descriptor, Part of PK |
NO |
- |
| Descriptor |
nvarchar (200) |
The DescriptorDefault value: '’ |
NO |
- |
| URL |
varchar (500) |
URL of the Descriptor. In case of a module related Descriptor, the link to the module entry resp. the related webserviceDefault value: '’ |
YES |
- |
| DescriptorType |
nvarchar (50) |
Type of the Descriptor as described in table CollectionEventSeriesDescriptorType_EnumDefault value: N’Descriptor’ |
YES |
Refers to table CollEventSeriesDescriptorType_Enum |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data.Default value: suser_sname() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when the data were first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data.Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when the data were last updated.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionEventSeries
- CollEventSeriesDescriptorType_Enum
Table CollectionEventSeriesImage
The images showing the sites of a series of collection events, e.g. an expedition
| Column |
Data type |
Description |
Nullable |
Relation |
| SeriesID |
int |
Unique ID for the table CollectionEventSeries (= foreign key and part of primary key) |
NO |
Refers to table CollectionEventSeries |
| URI |
varchar (255) |
The complete URI address of the image. This is only a cached value, if ResourceID is available and referring to the module DiversityResources |
NO |
- |
| ResourceURI |
varchar (255) |
The URI of the resource (e.g. see module DiversityResources) |
YES |
- |
| ImageType |
nvarchar (50) |
Type of the image, e.g. map |
YES |
Refers to table CollEventSeriesImageType_Enum |
| Notes |
nvarchar (MAX) |
Notes to this image of the collection site |
YES |
- |
| Description |
xml (MAX) |
Description of the image |
YES |
- |
| Title |
nvarchar (500) |
Title of the resource |
YES |
- |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
- |
| CreatorAgent |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
- |
| CreatorAgentURI |
varchar (255) |
Link to the module DiversityAgents |
YES |
- |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
- |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e.g. Creative Common licenses |
YES |
- |
| InternalNotes |
nvarchar (500) |
Internal notes which should not be published e.g. on websites |
YES |
- |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
- |
| LicenseHolderAgentURI |
nvarchar (500) |
The link to a module containing futher information on the person or institution holding the license |
YES |
- |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionEventSeries
- CollEventSeriesImageType_Enum
Table CollectionExternalDatasource
CollectionExternalDatasource document the sources of the names.
| Column |
Data type |
Description |
Nullable |
Relation |
| ExternalDatasourceID |
int |
An ID to identify an external data collection of the collected specimen (primary key, the ID has no meaning outside of the DiversityWorkbench system) |
NO |
- |
| ExternalDatasourceName |
nvarchar (255) |
The name of the data collection which has been integrated or can be linked to for further analysis |
YES |
- |
| ExternalDatasourceVersion |
nvarchar (255) |
The version of this data collection (either official version number or dates when the collection was integrated) |
YES |
- |
| Rights |
nvarchar (500) |
A description of copyright agreements or permission to use data from the external database |
YES |
- |
| ExternalDatasourceAuthors |
nvarchar (200) |
The persons or institutions responsible for the external database |
YES |
- |
| ExternalDatasourceURI |
nvarchar (300) |
The URI of the database provider or the external database |
YES |
- |
| ExternalDatasourceInstitution |
nvarchar (300) |
The institution responsible for the external database |
YES |
- |
| InternalNotes |
nvarchar (1500) |
Additional notes on this data collection |
YES |
- |
| ExternalAttribute_NameID |
nvarchar (255) |
The table and field name in the external data collection to which CollectionExternalID refers |
YES |
- |
| PreferredSequence |
tinyint |
For selection in e.g. picklists: of several equal names only the name from the source with the lowest preferred sequence will be provided. |
YES |
- |
| Disabled |
bit |
If this source should be disabled for selection of names e.g. in picklists |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Table CollectionImage
The images showing the collection
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionID |
int |
Refers to the ID of Collection (= foreign key and part of primary key) |
NO |
Refers to table Collection |
| URI |
varchar (255) |
The complete URI address of the image. |
NO |
- |
| ImageType |
nvarchar (50) |
Type of the image, e.g. label |
YES |
Refers to table CollCollectionImageType_Enum |
| Notes |
nvarchar (MAX) |
Notes on the collection image |
YES |
- |
| Description |
xml (MAX) |
Description of the image |
YES |
- |
| Title |
nvarchar (500) |
Title of the resource |
YES |
- |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
- |
| CreatorAgent |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
- |
| CreatorAgentURI |
varchar (255) |
Link to the module DiversityAgents |
YES |
- |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
- |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e.g. Creative Common licenses |
YES |
- |
| InternalNotes |
nvarchar (500) |
Internal notes which should not be published e.g. on websites |
YES |
- |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
- |
| LicenseHolderAgentURI |
nvarchar (500) |
The link to a module containing futher information on the person or institution holding the license |
YES |
- |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
- |
| LocationGeometry |
geometry (MAX) |
Geometry of the collection e.g. within a floor plan |
YES |
- |
| RecordingDate |
datetime |
The recording date of the resource |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| LogInsertedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollCollectionImageType_Enum
- Collection
Table CollectionManager
Collection managers within DiversityCollection responsible for specimen transactions
| Column |
Data type |
Description |
Nullable |
Relation |
| LoginName |
nvarchar (50) |
A login name which the user uses to access the DivesityWorkbench, Microsoft domains, etc.. |
NO |
- |
| AdministratingCollectionID |
int |
ID for the collection for which the Manager has the right to administrate the transaction. Corresponds to AdministratingCollectionID in table Transaction. |
NO |
Refers to table Collection |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table CollectionProject
The projects within which the collection specimen were placed
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimen |
| ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
NO |
Refers to table ProjectProxy |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimen
- ProjectProxy
trgInsCollectionProject
Setting LastChanges in table ProjectProxy
Table CollectionSpecimen
The data directly attributed to the collected specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Unique ID for the table CollectionSpecimen (primary key) |
NO |
- |
| Version |
int |
The version of the data setDefault value: (1) |
NO |
- |
| CollectionEventID |
int |
Refers to the ID of table CollectionEvent (= foreign key and part of primary key) |
YES |
Refers to table CollectionEvent |
| AccessionNumber |
nvarchar (50) |
Accession number of the specimen within the collection, e.g. “M-29834752” |
YES |
- |
| AccessionDate |
datetime |
The date of the accession calculated from the entries in AccessionDay, -Month and -Year |
YES |
- |
| AccessionDay |
tinyint |
The day of the date when the specimen was acquired in the collection |
YES |
- |
| AccessionMonth |
tinyint |
The month of the date when the specimen was acquired in the collection |
YES |
- |
| AccessionYear |
smallint |
The year of the date when the specimen was acquired in the collection |
YES |
- |
| AccessionDateSupplement |
nvarchar (255) |
Verbal or additional accession date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
YES |
- |
| AccessionDateCategory |
nvarchar (50) |
Category of the date of the accession e.g. “system”, “estimated” (= foreign key, see in table CollDateCategory_Enum) |
YES |
Refers to table CollDateCategory_Enum |
| DepositorsName |
nvarchar (255) |
The name of the depositor(s) (person or organization responsible for deposition). Where entire collections are deposited, this should also contain the collection name (e.g. ‘Herbarium P. Döbbler’) |
YES |
- |
| DepositorsAgentURI |
varchar (255) |
The URI of the depositor(s) (person or organization responsible for deposition) |
YES |
- |
| DepositorsAccessionNumber |
nvarchar (50) |
Accession number of the specimen within the previous or original collection, e.g. ‘D-23948’ |
YES |
- |
| LabelTitle |
nvarchar (MAX) |
The title of the label e.g. for printing labels. |
YES |
- |
| LabelType |
nvarchar (50) |
Printed, typewritten, typewritten with handwriting added, entirely in handwriting, etc. |
YES |
Refers to table CollLabelType_Enum |
| LabelTranscriptionState |
nvarchar (50) |
The state of the transcription of a label into the database: ‘Not started’, ‘incomplete’, ‘complete’ |
YES |
Refers to table CollLabelTranscriptionState_Enum |
| LabelTranscriptionNotes |
nvarchar (MAX) |
User defined notes on the transcription of the label into the database |
YES |
- |
| ExsiccataURI |
varchar (255) |
If specimen is an exsiccata: The URI of the exsiccata series, e.g. as stored within the DiversityExsiccata module |
YES |
- |
| ExsiccataAbbreviation |
nvarchar (255) |
If specimen is an exsiccata: Standard abbreviation of the exsiccata (not necessarily a unique identifier; editors or publication places may change over time) |
YES |
- |
| OriginalNotes |
nvarchar (MAX) |
Notes found on the label of the specimen by the original collector or from a later revision |
YES |
- |
| AdditionalNotes |
nvarchar (MAX) |
Additional notes made by the editor of the specimen record, e.g. ‘doubtful identification/locality’ |
YES |
- |
| Problems |
nvarchar (255) |
Description of a problem which occurred during data editing. Typically these entries should be deleted after help has been obtained. Do not enter scientific problems here; use AdditionalNotes for such permanent problems! |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise nullDefault value: N’Withhold by default’ |
YES |
- |
| InternalNotes |
nvarchar (MAX) |
Internal notes which should not be published e.g. on websites |
YES |
- |
| ExternalDatasourceID |
int |
An ID to identify an external data collection of the collected specimen (primary key, the ID has no meaning outside of the DiversityWorkbench system) |
YES |
Refers to table CollectionExternalDatasource |
| ExternalIdentifier |
nvarchar (100) |
The identifier of the external specimen as defined in the external data source |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollDateCategory_Enum
- CollectionEvent
- CollectionExternalDatasource
- CollLabelTranscriptionState_Enum
- CollLabelType_Enum
Table CollectionSpecimenImage
The images of a collection specimen or of an organism (stored in table IdentificationUnit) within this specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimen and table IdentificationUnit |
| URI |
varchar (255) |
The complete URI address of the image. This is only a cached value, if ResourceID is available and referring to the module DiversityResources |
NO |
- |
| ResourceURI |
varchar (255) |
The URI of the image, e.g. as stored in the module DiversityResources. |
YES |
- |
| SpecimenPartID |
int |
Optional: If the data set is not related to a part of a specimen, the ID of a related part (= foreign key) |
YES |
Refers to table CollectionSpecimenPart |
| IdentificationUnitID |
int |
If image refers to only one out of several IdentificationUnits for a specimen, refers to the ID of an IdentificationUnit for a collection specimen (= foreign key) |
YES |
Refers to table IdentificationUnit |
| ImageType |
nvarchar (50) |
Type of the image, e.g. photograph |
YES |
Refers to table CollSpecimenImageType_Enum |
| Notes |
nvarchar (MAX) |
Notes on the specimen image |
YES |
- |
| Description |
xml (MAX) |
Description of the image |
YES |
- |
| Title |
nvarchar (500) |
Title of the resource |
YES |
- |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
- |
| CreatorAgent |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
- |
| CreatorAgentURI |
varchar (255) |
Link to the module DiversityAgents |
YES |
- |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
- |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e. g. Creative Common Licenses |
YES |
- |
| LicenseURI |
varchar (500) |
The URI of the license for the resource |
YES |
- |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
- |
| LicenseHolderAgentURI |
nvarchar (500) |
The link to a module containing futher information on the person or institution holding the license |
YES |
- |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
- |
| LicenseNotes |
nvarchar (500) |
Notice on license for the resource |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| DisplayOrder |
int |
The order in which the images should be shown in a interface |
YES |
- |
| InternalNotes |
nvarchar (500) |
Internal notes which should not be published e.g. on websites |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimen
- CollectionSpecimenPart
- CollSpecimenImageType_Enum
- IdentificationUnit
CollectionSpecimenImage_URI
Deprecated
trgInsCollectionSpecimenImage
Setting the version of the dataset
Table CollectionSpecimenImageProperty
The properties of images of a collection specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenImage |
| URI |
varchar (255) |
The complete URI address of the image. This is only a cached value, if ResourceID is available and referring to the module DiversityResources |
NO |
Refers to table CollectionSpecimenImage |
| Property |
varchar (255) |
The property of the image |
NO |
- |
| Description |
nvarchar (MAX) |
If description of the property of the image |
YES |
- |
| ImageArea |
geometry (MAX) |
The area in the image the property refers to |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
| PropertyURI |
varchar (500) |
The URI of the property of the image, e.g. a link to module DiversityScientificTerms |
YES |
- |
Depending on:
Table CollectionSpecimenPart
Parts of a collected specimen. Includes a possible hierarchy of the parts
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimen and table CollectionSpecimenPart |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen (= part of primary key). |
NO |
- |
| DerivedFromSpecimenPartID |
int |
SpecimenPartID of the specimen from which the current specimen is derived from |
YES |
Refers to table CollectionSpecimenPart |
| PreparationMethod |
nvarchar (MAX) |
The method used for the preparation of the part of the specimen, e.g. the inoculation method for cultures |
YES |
- |
| PreparationDate |
datetime |
Point in time when the part was preparated e.g when it was separated from the source object |
YES |
- |
| AccessionNumber |
nvarchar (50) |
Accession number of the part of the specimen within the collection, if it is different from the accession number of the specimen as stored in the table CollectionSpecimen, e.g. “M-29834752” |
YES |
- |
| PartSublabel |
nvarchar (50) |
The label for a part of a specimen, e.g. “cone”, or a number attached to a duplicate of a specimen |
YES |
- |
| CollectionID |
int |
ID of the collection as stored in table Collection (= foreign key, see table Collection) |
NO |
Refers to table Collection |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. (= foreign key, see table CollMaterialCategory_Enum)Default value: N’specimen’ |
NO |
Refers to table CollMaterialCategory_Enum |
| StorageLocation |
nvarchar (255) |
A code identifying the place where the specimen is stored within the collection. Frequently the accepted scientific name is used as storage location code. |
YES |
- |
| Stock |
float |
Number of stock units, if the specimen is stored in separated units e.g. several boxes or vessels (max. 255) |
YES |
- |
| StockUnit |
nvarchar (50) |
If empty, the stock is given as a count, else it contains the unit in which stock is expressed, e.g. µl, ml, kg etc. |
YES |
- |
| StorageContainer |
nvarchar (500) |
The container in which the part is stored |
YES |
- |
| ResponsibleName |
nvarchar (255) |
Name of the person or institution responsible for the preparation |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or institution responsible for the preparation (= foreign key) as stored in the module DiversityAgents |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the storage of the sample |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the specimen part is withhold, the reason for withholding the data, otherwise null. |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- Collection
- CollectionSpecimen
- CollMaterialCategory_Enum
trgInsCollectionSpecimenPart
Setting the version of the dataset
Table CollectionSpecimenPartDescription
Description of the specimen part with a standardized vocabulary as defined in the module DiversityScientificTerms
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenPart |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen (= part of primary key). |
NO |
Refers to table CollectionSpecimenPart |
| PartDescriptionID |
int |
ID of the description (Part of primary key) |
NO |
- |
| IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
YES |
- |
| Description |
nvarchar (MAX) |
The descrition of the part. Cached value if DescriptionTermURI is used |
YES |
- |
| DescriptionTermURI |
varchar (500) |
Link to a external datasource like a webservice or the module DiversityScientificTerms where the description is documented |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes about this description |
YES |
- |
| DescriptionHierarchyCache |
nvarchar (MAX) |
Hierarchy of the description. For values linked to a module, a cached value provided by the module |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table CollectionSpecimenProcessing
The processing which was applied to a collected specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimen and table CollectionSpecimenPart |
| SpecimenProcessingID |
int |
Unique ID of the processing of a specimen or part of a specimen, part of primary key |
NO |
- |
| ProcessingDate |
datetime |
Point in time of the start of the processing |
YES |
- |
| ProcessingID |
int |
ID of the processing method. Refers to ProcessingID in table Processing (foreign key)Default value: (1) |
NO |
Refers to table Processing |
| Protocoll |
nvarchar (100) |
The label of the processing protocol |
YES |
- |
| SpecimenPartID |
int |
Optional: If the data set is related to a part of a specimen, the ID of a related part (= foreign key, see table CollectionSpecimenPart) |
YES |
Refers to table CollectionSpecimenPart |
| ProcessingDuration |
varchar (50) |
The duration of the processing including the unit (e.g. 5 min) or the end of the processing starting at the processing date (e.g. 23.05.2008) |
YES |
- |
| ResponsibleName |
nvarchar (255) |
Name of the person or institution responsible for the determination |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the processing |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimen
- CollectionSpecimenPart
- Processing
trgInsCollectionSpecimenProcessing
Setting the version of the dataset
Table CollectionSpecimenProcessingMethod
The methods used for a processing of a specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to ID of CollectionSpecimen (= Foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenProcessing |
| SpecimenProcessingID |
int |
Refers to the ID of the specimen processing (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenProcessing |
| MethodID |
int |
ID of the method, part of primary key |
NO |
Refers to table MethodForProcessing |
| MethodMarker |
nvarchar (50) |
A marker for the method, part of primary keyDefault value: ‘1’ |
NO |
- |
| ProcessingID |
int |
ID of the processing. Refers to ProcessingID in table Processing (foreign key)Default value: (1) |
NO |
Refers to table MethodForProcessing |
| LogCreatedWhen |
datetime |
The time when this dataset was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Who created this datasetDefault value: user_name() |
YES |
- |
| LogUpdatedWhen |
datetime |
The last time when this dataset was updatedDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Who was the last to update this datasetDefault value: user_name() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimenProcessing
- MethodForProcessing
Table CollectionSpecimenProcessingMethodParameter
The parameter values of a method used for the processing of a specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to ID of CollectionSpecimen (= Foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenProcessingMethod |
| SpecimenProcessingID |
int |
Refers to ID of CollectionSpecimenProcessing (= Foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenProcessingMethod |
| ProcessingID |
int |
ID of the processing. Refers to ProcessingID in table Processing (= Foreign key and part of primary key)Default value: (1) |
NO |
Refers to table CollectionSpecimenProcessingMethod |
| MethodID |
int |
ID of the method (= Foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenProcessingMethod and table Parameter |
| MethodMarker |
nvarchar (50) |
A marker for the method, part of primary keyDefault value: ‘1’ |
NO |
Refers to table CollectionSpecimenProcessingMethod |
| ParameterID |
int |
ID of the parameter. Referes to table Parameter (= Foreign key and part of primary key) |
NO |
Refers to table Parameter |
| Value |
nvarchar (MAX) |
The value of the parameter if different of the default value as documented in the table Parameter |
NO |
- |
| LogCreatedWhen |
datetime |
The time when this dataset was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Who created this datasetDefault value: user_name() |
YES |
- |
| LogUpdatedWhen |
datetime |
The last time when this dataset was updatedDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Who was the last to update this datasetDefault value: user_name() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimenProcessingMethod
- Parameter
Table CollectionSpecimenReference
A reference related to the collection specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to unique ID of collection specimen record (part of primary key) |
NO |
Refers to table CollectionSpecimen and table IdentificationUnit |
| ReferenceID |
int |
Unique reference ID for the reference record (part of primary key) |
NO |
- |
| ReferenceTitle |
nvarchar (400) |
The title of the publication related to the specimen or parts of it. Note that this is only a cached value where ReferenceURI is present |
NO |
- |
| ReferenceURI |
varchar (500) |
URI of the reference, e.g. a connection to the module DiversityReferences |
YES |
- |
| IdentificationUnitID |
int |
If relation refers to a certain organism within a specimen, the ID of an IdentificationUnit (= foreign key) |
YES |
Refers to table IdentificationUnit |
| IdentificationSequence |
smallint |
Referes to table Identification: The sequence of the identifications. |
YES |
- |
| SpecimenPartID |
int |
If the relation refers to a part of a specimen, the ID of a related part (= foreign key) |
YES |
Refers to table CollectionSpecimenPart |
| ReferenceDetails |
nvarchar (500) |
The exact location within the reference, e.g. pages, plates |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes about the reference |
YES |
- |
| ResponsibleName |
nvarchar (255) |
The name of the agent (person or organization) responsible for this entry. |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or organisation responsible for the data (see e.g. module DiversityAgents) |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimen
- CollectionSpecimenPart
- IdentificationUnit
Table CollectionSpecimenRelation
The relations of a collection specimen to other collection specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Unique reference ID for the collection specimen record (primary key) |
NO |
Refers to table CollectionSpecimen and table IdentificationUnit |
| RelatedSpecimenURI |
varchar (255) |
URI of the related specimen |
NO |
- |
| RelatedSpecimenDisplayText |
varchar (255) |
The name of a related specimen as shown e.g. in a user interface |
NO |
- |
| RelationType |
nvarchar (50) |
Type of the relation between the specimen (= foreign key, see table CollRelationType_Enum) |
YES |
Refers to table CollSpecimenRelationType_Enum |
| RelatedSpecimenCollectionID |
int |
ID of the Collection as stored in table Collection (= foreign key, see table Collection) |
YES |
Refers to table Collection |
| RelatedSpecimenDescription |
nvarchar (MAX) |
Description of the related specimen |
YES |
- |
| IdentificationUnitID |
int |
If relation refers to a certain organism within a specimen, the ID of an IdentificationUnit (= foreign key) |
YES |
Refers to table IdentificationUnit |
| SpecimenPartID |
int |
If the relation refers to a part of a specimen, the ID of a related part (= foreign key) |
YES |
Refers to table CollectionSpecimenPart |
| Notes |
nvarchar (MAX) |
Notes on the relation to the specimen |
YES |
- |
| IsInternalRelationCache |
bit |
If the relation represents a connection between specimen in this databaseDefault value: (1) |
NO |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- Collection
- CollectionSpecimen
- CollectionSpecimenPart
- CollSpecimenRelationType_Enum
- IdentificationUnit
trgInsCollectionSpecimenRelation
Setting the version of the dataset
Table CollectionSpecimenTransaction
The transactions in which a specimen was involved
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenPart |
| TransactionID |
int |
Unique ID for the table Transaction (= foreign key and part of primary key) |
NO |
Refers to table Transaction |
| SpecimenPartID |
int |
Optional: If the data set is related to a part of a specimen, the ID of a related part (= foreign key, see table CollectionSpecimenPart) |
NO |
Refers to table CollectionSpecimenPart |
| AccessionNumber |
nvarchar (255) |
Accession number which has been assigen to the part of the specimen, e.g. in connection with a former inventory. |
YES |
- |
| TransactionReturnID |
int |
Unique ID for the table Transaction (= foreign key) for the return of a part that has been on loan |
YES |
- |
| TransactionTitle |
nvarchar (200) |
Title as in related table Transaction. Used for validation of correct entry of transaction with type regulation (see insert trigger) |
YES |
- |
| IsOnLoan |
bit |
True, if a specimen is on loan |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user to first enter (typ or import) the data.Default value: suser_sname() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Point in time when the data was first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last.Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Point in time when this data set was updated last.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimenPart
- Transaction
trgInsCollectionSpecimenTransaction
Rollback of regulations if corresponding datasets are missing in table CollectionEventRegulation
Table CollectionTask
A task for a collection. Details are defined in table Task
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionTaskID |
int |
PK of the table |
NO |
- |
| CollectionTaskParentID |
int |
Relation to PK for hierarchy within the data |
YES |
Refers to table CollectionTask |
| CollectionID |
int |
Relation to table Collection. Every ColletionTask needs a relation to a collection |
NO |
Refers to table Collection |
| TaskID |
int |
Relation to table Task where details for the collection task are defined |
NO |
Refers to table Task |
| DisplayOrder |
int |
Display order e.g. in a report. Data with value 0 will not be includedDefault value: (1) |
YES |
- |
| DisplayText |
nvarchar (400) |
The display text of the module related data as shown e.g. in a user interface |
YES |
- |
| CollectionSpecimenID |
int |
Refers to the CollectionSpecimenID of CollectionSpecimenPart (= foreign key and part of primary key) |
YES |
Refers to table CollectionSpecimenPart |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen the task is related to. |
YES |
Refers to table CollectionSpecimenPart |
| TransactionID |
int |
ID of a transaction. Related to PK of table Transaction |
YES |
Refers to table Transaction |
| ModuleUri |
varchar (500) |
The URL of module related data |
YES |
- |
| TaskStart |
datetime |
The start date and or time of the collection tasks. The type is defined in table Task |
YES |
- |
| TaskEnd |
datetime |
The end date and or time of the collection tasks. The type is defined in table Task |
YES |
- |
| Result |
nvarchar (400) |
A text result either taken from a list or entered. The type is defined in table Task |
YES |
- |
| URI |
varchar (500) |
The URI of the collection tasks. The type is defined in table Task |
YES |
- |
| NumberValue |
real |
The numeric value of the collection tasks. The type is defined in table Task |
YES |
- |
| BoolValue |
bit |
The boolean of the collection tasks. The type is defined in table Task |
YES |
- |
| MetricDescription |
nvarchar (500) |
Description of the metric e.g. imported from a time series database like Prometheus |
YES |
- |
| MetricSource |
varchar (4000) |
The source of the metric e.g. the PromQL statement for the import from a timeseries database Prometheus |
YES |
- |
| MetricUnit |
nvarchar (50) |
The unit of the metric, e.g. °C for temperature |
YES |
- |
| ResponsibleAgent |
nvarchar (500) |
The name of the responsible person or institution |
YES |
- |
| ResponsibleAgentURI |
varchar (500) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
YES |
- |
| Description |
nvarchar (MAX) |
The description of the collection tasks. The type is defined in table Task |
YES |
- |
| Notes |
nvarchar (MAX) |
The notes of the collection tasks. The type is defined in table Task |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: dbo.UserID() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: dbo.UserID() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- Collection
- CollectionSpecimenPart
- Task
- Transaction
trgInsCollectionTask
Inserting CollectionID of parent entry if missing
Table CollectionTaskImage
The images showing the CollectionTask
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionTaskID |
int |
Refers to the ID of CollectionTask (= foreign key and part of primary key) |
NO |
Refers to table CollectionTask |
| URI |
varchar (255) |
The complete URI address of the image. |
NO |
- |
| ImageType |
nvarchar (50) |
Type of the image, e.g. label |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the CollectionTask image |
YES |
- |
| Description |
xml (MAX) |
Description of the image |
YES |
- |
| Title |
nvarchar (500) |
Title of the resource |
YES |
- |
| ObjectGeometry |
geometry (MAX) |
The geometry of an object placed within the image |
YES |
- |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
- |
| CreatorAgent |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
- |
| CreatorAgentURI |
varchar (255) |
Link to the module DiversityAgents |
YES |
- |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
- |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e.g. Creative Common licenses |
YES |
- |
| InternalNotes |
nvarchar (500) |
Internal notes which should not be published e.g. on websites |
YES |
- |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
- |
| LicenseHolderAgentURI |
nvarchar (500) |
The link to a module containing futher information on the person or institution holding the license |
YES |
- |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
- |
| DisplayOrder |
smallint |
The display order of the image |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| LogInsertedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: dbo.UserID() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: dbo.UserID() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table CollectionTaskMetric
The metric related to a collection task
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionTaskID |
int |
Refers to the ID of CollectionTask (= foreign key and part of primary key) |
NO |
Refers to table CollectionTask |
| MetricDate |
datetime |
Date and time of the metric, part of PK |
NO |
- |
| Aggregation |
nvarchar (50) |
The Aggregation applied for retrieval of the value, e.g. max, avg etc.Default value: N’none’ |
NO |
Refers to table CollTaskMetricAggregation_Enum |
| MetricValue |
real |
The value of the metric |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: dbo.UserID() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: dbo.UserID() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionTask
- CollTaskMetricAggregation_Enum
Table CollectionUser
Users of collections within DiversityCollection
| Column |
Data type |
Description |
Nullable |
Relation |
| LoginName |
nvarchar (50) |
A login name which the user uses to access the DivesityWorkbench, Microsoft domains, etc.. |
NO |
- |
| CollectionID |
int |
ID for the collection for the user has access to administrate the transaction. |
NO |
Refers to table Collection |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table Entity
The entities in an application e.g. the tables and columns in a database
| Column |
Data type |
Description |
Nullable |
Relation |
| Entity |
varchar (500) |
The name of the entity, e.g. Table.Column.Content within the database or a unique string for e.g. a message within the DiversityWorkbench e.g. “DiversityWorkbench.Message.Connection.NoAccess”, PK |
NO |
- |
| DisplayGroup |
nvarchar (50) |
If DiversityWorkbench entities should be displayed in a group, the name of the group |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the entity |
YES |
- |
| Obsolete |
bit |
True if an entity is obsolete. Obsolete entities may be kept to ensure compatibility with older modules |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Table EntityRepresentation
The description of the entity in a certain context in different languages
| Column |
Data type |
Description |
Nullable |
Relation |
| Entity |
varchar (500) |
The name of the entity. Foreign key, relates to table Entity |
NO |
Refers to table Entity |
| LanguageCode |
nvarchar (50) |
ISO 639: 2-letter codes for the language of the content |
NO |
Refers to table EntityLanguageCode_Enum |
| EntityContext |
nvarchar (50) |
The context for the representation, e.g. “Exchange with ABCD”, “collection management” or “observation” as defined in table EntityContext_Enum |
NO |
Refers to table EntityContext_Enum |
| DisplayText |
nvarchar (50) |
The text for the entity as shown e.g. in a user interface |
YES |
- |
| Abbreviation |
nvarchar (20) |
The abbreviation for the entity as shown e.g. in a user interface |
YES |
- |
| Description |
nvarchar (MAX) |
The description of the entity |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the representation of the entity |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- Entity
- EntityContext_Enum
- EntityLanguageCode_Enum
Table EntityUsage
The usage of an entity in a certain context, e.g. hidden, readonly
| Column |
Data type |
Description |
Nullable |
Relation |
| Entity |
varchar (500) |
The name of the entity. Foreign key, relates to table Entity |
NO |
Refers to table Entity |
| EntityContext |
nvarchar (50) |
The context for the representation, e.g. “Exchange with ABCD”, “collection management” or “observation” as defined in table EntityContext_Enum |
NO |
Refers to table EntityContext_Enum |
| Accessibility |
nvarchar (50) |
If the access of entity is resticted to e.g. read only or it can be edited without restrictions |
YES |
Refers to table EntityAccessibility_Enum |
| Determination |
nvarchar (50) |
If a value is determined e.g. by the system or the user |
YES |
Refers to table EntityDetermination_Enum |
| Visibility |
nvarchar (50) |
If the entity is visible or hidden from e.g. a user interface |
YES |
Refers to table EntityVisibility_Enum |
| PresetValue |
nvarchar (500) |
If a value is preset, the value or SQL statement for the value, e.g. ‘determination’ for identifications when using a mobile device during an expedition |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the usage of the entity |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- Entity
- EntityAccessibility_Enum
- EntityContext_Enum
- EntityDetermination_Enum
- EntityVisibility_Enum
Table ExternalIdentifier
An external identier related to a dataset, e.g. a DOI
| Column |
Data type |
Description |
Nullable |
Relation |
| ID |
int |
ID of the identifier (Primary key) |
NO |
- |
| ReferencedTable |
nvarchar (128) |
The name of the table the external identifier refers to |
NO |
- |
| ReferencedID |
int |
The ID of the data set in the table the external identifier refers to |
NO |
- |
| Type |
nvarchar (50) |
The type of the identifier as defined in table ExternalIdentifierType |
YES |
Refers to table ExternalIdentifierType |
| Identifier |
nvarchar (500) |
The identifier |
YES |
- |
| URL |
varchar (500) |
A URL with further informations about the identifier |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes about the identifier |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table ExternalIdentifierType
The type of an external identier, e.g. DOI
| Column |
Data type |
Description |
Nullable |
Relation |
| Type |
nvarchar (50) |
The type of external identifiers (primary key) |
NO |
- |
| ParentType |
nvarchar (50) |
The superior type of this type |
YES |
- |
| URL |
varchar (500) |
A URL providing further informations about this type |
YES |
- |
| Description |
nvarchar (MAX) |
The description of this type |
YES |
- |
| InternalNotes |
nvarchar (MAX) |
Internal notes about the type |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newid() |
NO |
- |
Table Identification
The identifications of the organisms within a specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table Identification and table IdentificationUnit |
| IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
NO |
Refers to table Identification and table IdentificationUnit |
| IdentificationSequence |
smallint |
The sequence of the identifications. The last identification (having the highest sequence) is regarded as validDefault value: (1) |
NO |
- |
| DependsOnIdentificationSequence |
smallint |
if the identification depends on another identification, e.g. for rocks where several terms from a terminology should be included |
YES |
Refers to table Identification |
| IdentificationDate |
datetime |
The date of the identification calculated from the entries in IdentificationDay, -Month and -Year |
YES |
- |
| IdentificationDay |
tinyint |
The day of the identification |
YES |
- |
| IdentificationMonth |
tinyint |
The month of the identification |
YES |
- |
| IdentificationYear |
smallint |
The year of the identification. The year may be empty if only the day or month are known. |
YES |
- |
| IdentificationDateSupplement |
nvarchar (255) |
Verbal or additional identification date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
YES |
- |
| IdentificationDateCategory |
nvarchar (50) |
Category of the date of the identification e.g. “system”, “estimated” (= foreign key, see in table CollDateCategory_Enum) |
YES |
Refers to table CollIdentificationDateCategory_Enum |
| VernacularTerm |
nvarchar (255) |
Name or term other than a taxonomic (= scientific) name, e.g. ‘pine’, ’limestone’, ‘conifer’, ‘hardwood’ |
YES |
- |
| TermURI |
varchar (500) |
The URI of the term, e.g. as provided by the module DiversityScientificTerms. |
YES |
- |
| TaxonomicName |
nvarchar (255) |
Valid name of the species (including the taxonomic author where available). Example: ‘Rosa canina L.’ |
YES |
- |
| NameURI |
varchar (255) |
The URI of the taxonomic name, e.g. as provided by the module DiversityTaxonNames. |
YES |
- |
| IdentificationCategory |
nvarchar (50) |
Category of the identification e.g. ‘determination’, ‘confirmation’, ‘absence’ (= foreign key, see table CollIdentificationCategory_Enum) |
YES |
Refers to table CollIdentificationCategory_Enum |
| IdentificationQualifier |
nvarchar (50) |
Qualification of the identification e.g. “cf.”," aff.", “sp. nov.” (= foreign key, see table CollIdentificationQualifier_Enum) |
YES |
Refers to table CollIdentificationQualifier_Enum |
| TypeStatus |
nvarchar (50) |
If identification unit is type of a taxonomic name: holotype, syntype, etc. (= foreign key, see table CollTypeStatus_Enum) |
YES |
Refers to table CollTypeStatus_Enum |
| TypeNotes |
nvarchar (MAX) |
Notes on the typification of this specimen |
YES |
- |
| Notes |
nvarchar (MAX) |
User defined notes, e.g. the reason for a re-determination / change of the name, etc. |
YES |
- |
| ResponsibleName |
nvarchar (255) |
Name of the person or institution responsible for the determination |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollIdentificationCategory_Enum
- CollIdentificationDateCategory_Enum
- CollIdentificationQualifier_Enum
- CollTypeStatus_Enum
- IdentificationUnit
trgIdentificationInsert
Updating the LastIdentificationCache in IdentificationUnit
trgInsIdentification
Updating empty date columns depending on a given date
Table IdentificationUnit
Organism which is present in or on a collectied specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimen and table IdentificationUnit |
| IdentificationUnitID |
int |
ID of the IdentificationUnit (= part of primary key). Usually one of possibly several organisms present on the collected specimen. Example: parasite with hyperparasite on plant leaf = 3 units, |
NO |
- |
| LastIdentificationCache |
nvarchar (255) |
The last identification as entered in table Identification |
NO |
- |
| FamilyCache |
nvarchar (255) |
A cached value of the family of the taxon of the last identification. Can be set by the editor, if NameURI in table Identification is NULL, otherwise set by the system. |
YES |
- |
| OrderCache |
nvarchar (255) |
A cached value of the order of the taxon of the last identification. Can be set by the editor, if NameURI in table Identification is NULL, otherwise set by the system. |
YES |
- |
| HierarchyCache |
nvarchar (500) |
A cached value fo the superior taxonomy of the last identification as derived from a taxonomic data provider |
YES |
- |
| TaxonomicGroup |
nvarchar (50) |
Taxonomic group the organism, identified by this unit, belongs to. Groups listed in table CollTaxonomicGroup_Enum (= foreign key) |
NO |
Refers to table CollTaxonomicGroup_Enum |
| OnlyObserved |
bit |
True, if the organism was only observed rather than collected. It is therefore not present on the preserved specimen. Example: Tree under which the collected mycorrhizal fungus grew.Default value: (0) |
YES |
- |
| RelatedUnitID |
int |
The IdentificationUnitID of the organism or substrate on which this organism is growing (= foreign key) |
YES |
Refers to table IdentificationUnit |
| RelationType |
nvarchar (50) |
The relation of a unit to its substrate, e.g. parasitism, symbiosis etc. as stored in CollRelationType_Enum (= foreign key) |
YES |
Refers to table CollUnitRelationType_Enum |
| ParentUnitID |
int |
The IdentificationUnitID of a parent organism of which this organism is a child of (= foreign key). |
YES |
Refers to table IdentificationUnit |
| ColonisedSubstratePart |
nvarchar (255) |
If a substrate association exists: part of the substrate which is affected in the interaction (e.g. ’leaves’, if a fungus is growing on the leaves of an infected plant) |
YES |
- |
| LifeStage |
nvarchar (255) |
Examples: ‘II, III’ for spore generations of rusts or ‘seed’, ‘seedling’ etc. for higher plants |
YES |
- |
| Gender |
nvarchar (50) |
The sex of the organism, e.g. ‘female’ |
YES |
- |
| NumberOfUnits |
smallint |
The number of units of this organism, e.g. 400 beetles in a bottle |
YES |
- |
| NumberOfUnitsModifier |
nvarchar (100) |
A modifier for the number of units of this organism, e.g. ca. 400 beetles in a bottle |
YES |
- |
| ExsiccataNumber |
nvarchar (50) |
If specimen is an exsiccata: Number of current specimen within the exsiccata series |
YES |
- |
| ExsiccataIdentification |
smallint |
Refers to the IdentificationSequence in Identification (= foreign key). The name under which the collectied specimen or this organism is published within an exsiccata. |
YES |
- |
| UnitIdentifier |
nvarchar (50) |
An identifier for the identification of the unit, e.g. a number painted on a tree within an experimental plot |
YES |
- |
| UnitDescription |
nvarchar (50) |
Description of the unit, especially if not an organism but parts or remnants of it were present or observed, e.g. a nest of an insect or a song of a bird |
YES |
- |
| Circumstances |
nvarchar (50) |
Circumstances of the occurence of the organism |
YES |
Refers to table CollCircumstances_Enum |
| RetrievalType |
nvarchar (50) |
The way the data about the unit were retrieved, e.g. observation, literature |
YES |
Refers to table CollRetrievalType_Enum |
| DisplayOrder |
smallint |
The sequence in which the units within this specimen will appear on e.g. a label where the first unit may be printed in the header and others in the text below. 0 means the unit should not appear on a label.Default value: (1) |
NO |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| Notes |
nvarchar (MAX) |
Further information on the organism or interaction, e.g. infection symptoms like ‘producing galls’ |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollCircumstances_Enum
- CollectionSpecimen
- CollRetrievalType_Enum
- CollTaxonomicGroup_Enum
- CollUnitRelationType_Enum
trgInsIdentificationUnit
setting the display oder for the new unit to the next number if none or an already present on was given
Table IdentificationUnitAnalysis
The analysis values taken from an organism resp. object
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table IdentificationUnit and table IdentificationUnitInPart |
| IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
NO |
Refers to table IdentificationUnit and table IdentificationUnitInPart |
| AnalysisID |
int |
Analysis ID, foreign key of table Analysis. |
NO |
Refers to table Analysis |
| AnalysisNumber |
nvarchar (50) |
Number of the analysis |
NO |
- |
| AnalysisResult |
nvarchar (MAX) |
The result of the analysis |
YES |
- |
| ExternalAnalysisURI |
varchar (255) |
An URI for an analysis as defined in an external datasoure |
YES |
- |
| ResponsibleName |
nvarchar (255) |
Name of the person or institution responsible for the determination |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
YES |
- |
| AnalysisDate |
nvarchar (50) |
The date of the analysis |
YES |
- |
| SpecimenPartID |
int |
ID of the part of a specimen (optional, foreign key) if the analysis was done with a part of the specimen (see table CollectionSpecimenPart). |
YES |
Refers to table IdentificationUnitInPart |
| Notes |
nvarchar (MAX) |
Notes on this analysis |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
| ToolUsage |
xml (MAX) |
The tools used for the analysis and their usage or settings. |
YES |
- |
Depending on:
- Analysis
- IdentificationUnit
- IdentificationUnitInPart
trgInsIdentificationUnitAnalysis
Setting the version of the dataset
Table IdentificationUnitAnalysisMethod
The methods used for an analysis
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to ID of CollectionSpecimen (= Foreign key and part of primary key) |
NO |
Refers to table IdentificationUnitAnalysis |
| IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
NO |
Refers to table IdentificationUnitAnalysis |
| MethodID |
int |
ID of the method, part of primary key |
NO |
Refers to table MethodForAnalysis |
| MethodMarker |
nvarchar (50) |
A marker for the method, part of primary keyDefault value: ‘1’ |
NO |
- |
| AnalysisID |
int |
ID of the processing. Refers to AnalysisID in table Processing (foreign key)Default value: (1) |
NO |
Refers to table IdentificationUnitAnalysis and table MethodForAnalysis |
| AnalysisNumber |
nvarchar (50) |
Number of the analysis |
NO |
Refers to table IdentificationUnitAnalysis |
| LogCreatedWhen |
datetime |
The time when this dataset was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Who created this datasetDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
The last time when this dataset was updatedDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Who was the last to update this datasetDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- IdentificationUnitAnalysis
- MethodForAnalysis
Table IdentificationUnitAnalysisMethodParameter
The parameter values of a method used for an analysis
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to ID of CollectionSpecimen (= Foreign key and part of primary key) |
NO |
Refers to table IdentificationUnitAnalysisMethod |
| IdentificationUnitID |
int |
ID of the identification unit (= Foreign key and part of primary key) |
NO |
Refers to table IdentificationUnitAnalysisMethod |
| AnalysisID |
int |
ID of the analysis. Refers to AnalysisID in table Analysis (= Foreign key and part of primary key)Default value: (1) |
NO |
Refers to table IdentificationUnitAnalysisMethod |
| AnalysisNumber |
nvarchar (50) |
Number of the analysis (= Foreign key and part of primary key) |
NO |
Refers to table IdentificationUnitAnalysisMethod |
| MethodID |
int |
ID of the method (= Foreign key and part of primary key) |
NO |
Refers to table IdentificationUnitAnalysisMethod and table Parameter |
| MethodMarker |
nvarchar (50) |
A marker for the method, part of primary keyDefault value: ‘1’ |
NO |
Refers to table IdentificationUnitAnalysisMethod |
| ParameterID |
int |
ID of the parameter tool. Referes to table Parameter (= Foreign key and part of primary key) |
NO |
Refers to table Parameter |
| Value |
nvarchar (MAX) |
The value of the parameter if different of the default value as documented in the table Parameter |
YES |
- |
| LogCreatedWhen |
datetime |
The time when this dataset was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Who created this datasetDefault value: user_name() |
YES |
- |
| LogUpdatedWhen |
datetime |
The last time when this dataset was updatedDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Who was the last to update this datasetDefault value: user_name() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- IdentificationUnitAnalysisMethod
- Parameter
Table IdentificationUnitGeoAnalysis
The geographical position or region of an organism at a certain time
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table IdentificationUnit |
| IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
NO |
Refers to table IdentificationUnit |
| AnalysisDate |
datetime |
The date of the analysis |
NO |
- |
| Geography |
geography |
The geography where the organism resp. object was located according to WGS84, e.g. a point (latitide, longitude and altitude) |
YES |
- |
| Geometry |
geometry (MAX) |
The geometry of the place the organism resp. object was observed, e.g. an area |
YES |
- |
| ResponsibleName |
nvarchar (255) |
Name of the person or institution responsible for the determination |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on this analysis |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table IdentificationUnitInPart
The list of the organisms which are found in a part of the specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenPart and table IdentificationUnit |
| IdentificationUnitID |
int |
ID of the identification unit in table IdentificationUnit (= part of primary key). Usually one of possibly several organisms present on the collection specimen. Example: parasite with hyperparasite on plant leaf = 3 units, |
NO |
Refers to table IdentificationUnit |
| SpecimenPartID |
int |
ID of the part of a specimen (optional, foreign key), if the identification unit is located on a part of the specimen (see table CollectionSpecimenPart). |
NO |
Refers to table CollectionSpecimenPart |
| DisplayOrder |
smallint |
The sequence in which the units within this part will appear on e.g. a label where the first unit may be printed in the header and others in the text below. 0 means the unit should not appear on a label.Default value: (1) |
NO |
- |
| LogInsertedBy |
nvarchar (50) |
Name of the user to first enter (typ or import) the data.Default value: suser_sname() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Point in time when the data was first entered (typed or imported) into this database.Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data last.Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Point in time when this data was updated last.Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimenPart
- IdentificationUnit
trgInsIdentificationUnitInPart
Setting the version of the dataset
Table LocalisationSystem
The geographic localisation systems, e.g. coordinates
| Column |
Data type |
Description |
Nullable |
Relation |
| LocalisationSystemID |
int |
Unique ID for the localisation system (= Primary key) |
NO |
- |
| LocalisationSystemParentID |
int |
LocalisationSystemID of the superior LocalisationSystem |
YES |
Refers to table LocalisationSystem |
| LocalisationSystemName |
nvarchar (100) |
Name of the system used for the determination of the place of the collection, e. g. Gauss-Krüger, MTB, GIS |
NO |
- |
| DefaultAccuracyOfLocalisation |
nvarchar (50) |
The default for the accuracy of values which can be reached with this method |
YES |
- |
| DefaultMeasurementUnit |
nvarchar (50) |
The default measurement unit for the localisation system, e.g. m, geograpic coordinates |
YES |
- |
| ParsingMethodName |
nvarchar (50) |
Internal value, specifying a programming method used for parsing text in fields Location1/Location2 in table CollectionLocalisation |
YES |
- |
| DisplayText |
nvarchar (50) |
Short abbreviated description of the localisation system as displayed in the user interface |
YES |
- |
| DisplayEnable |
bit |
Specifies, if this item is enabled to be used within the database. Localisation systems can be disabled to avoid seeing them, but keep the definition for the future. |
YES |
- |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
YES |
- |
| Description |
nvarchar (255) |
Description of the localisation method |
YES |
- |
| DisplayTextLocation1 |
nvarchar (50) |
Short abbreviated description of the attribute Location1 in the table CollectionGeography as displayed in the user interface |
YES |
- |
| DescriptionLocation1 |
nvarchar (255) |
Description of the attribute Location1 in the table CollectionGeography as displayed in the user interface |
YES |
- |
| DisplayTextLocation2 |
nvarchar (50) |
Short abbreviated description of the attribute Location2 in the table CollectionGeography as displayed in the user interface |
YES |
- |
| DescriptionLocation2 |
nvarchar (255) |
Description of the attribute Location2 in the table CollectionGeography as displayed in the user interface |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Content
This is the default content corresponding to the default settings in the client software.
| LocalisationSystemID |
DisplayText |
DescriptionLocation1 |
DescriptionLocation2 |
| 1 |
Top50 (deutsche Landesvermessung) - deprecated |
Place |
Coordinates (E, N) |
| 2 |
Gauss-Krüger coordinates |
R |
H |
| 3 |
MTB (A, CH, D) |
Messtischblatt |
Quadrant |
| 4 |
Altitude (mNN) |
Altitude from |
Altitude to |
| 5 |
mNN (barometric) |
Altitude from |
Altitude to |
| 6 |
Greenwich Coordinates |
Longitude (EW) |
Latitude (NS) |
| 7 |
Named area (DiversityGazetteers) |
Location |
Thesaurus code |
| 8 |
Coord. WGS84 |
Longitude (East-West) |
Latitude (North-South) |
| 9 |
Coordinates |
EW |
NS |
| 10 |
Exposition |
Exposition from |
Exposition to |
| 11 |
Slope |
Slope from |
Slope to |
| 12 |
Coord. PD |
Longitude (EW) |
Latitude (NS) |
| 13 |
Sampling plot (DiversitySamplingPlots) |
The name of the sampling plot. A cached value if the entry is linked to the module DiversitySamplingPlots |
The link to the module DiversitySamplingPlots |
| 14 |
Depth |
All material collected was at least this deep |
All material collected was at most this deep |
| 15 |
Height |
All material collected was at least at this hight |
All material collected was at most at this hight |
| 17 |
ÖK |
Österreichische Karte |
Quadrant |
| 18 |
2. Named area (DiversityGazetteers) |
Location |
Thesaurus code |
| 19 |
3. Named area (DiversityGazetteers) |
Location |
Thesaurus code |
| 20 |
4. Named area (DiversityGazetteers) |
Location |
Thesaurus code |
| 21 |
5. Named area (DiversityGazetteers) |
Location |
Thesaurus code |
| 22 |
UTM |
East |
North |
Table Method
Methods used within the database
| Column |
Data type |
Description |
Nullable |
Relation |
| MethodID |
int |
ID of the Method (Primary key) |
NO |
- |
| MethodParentID |
int |
MethodID of the parent Method, if it belongs to a certain type documented in this table |
YES |
Refers to table Method |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entriesDefault value: (0) |
YES |
- |
| DisplayText |
nvarchar (50) |
Name of the Method as e.g. shown in user interface |
YES |
- |
| Description |
nvarchar (MAX) |
Description of the Method |
YES |
- |
| MethodURI |
varchar (255) |
URI referring to an external documentation of the Method |
YES |
- |
| ForCollectionEvent |
bit |
If a method may be used during a collection event |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on this method |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Table MethodForAnalysis
Methods available for a Analysis
| Column |
Data type |
Description |
Nullable |
Relation |
| AnalysisID |
int |
ID of the table Analysis (foreign key and part of primary key) |
NO |
Refers to table Analysis |
| MethodID |
int |
ID of the table Method (foreign key and part of primary key) |
NO |
Refers to table Method |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table MethodForProcessing
Methods available for a processing
| Column |
Data type |
Description |
Nullable |
Relation |
| ProcessingID |
int |
ID of the table Processing (foreign key and part of primary key) |
NO |
Refers to table Processing |
| MethodID |
int |
ID of the table Method (foreign key and part of primary key) |
NO |
Refers to table Method |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table Parameter
The variable parameters within a method
| Column |
Data type |
Description |
Nullable |
Relation |
| MethodID |
int |
ID of the Method (foreign key and part of primary key) |
NO |
Refers to table Method |
| ParameterID |
int |
ID of the Parameter (part of primary key) |
NO |
- |
| DisplayText |
nvarchar (50) |
Name of the parameter as e.g. shown in user interface |
YES |
- |
| Description |
nvarchar (MAX) |
Description of the parameter |
YES |
- |
| ParameterURI |
varchar (255) |
URI referring to an external documentation of the Parameter |
YES |
- |
| DefaultValue |
nvarchar (MAX) |
The default value of the parameter |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on this parameter |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table Processing
The processings of the specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| ProcessingID |
int |
ID of the processing (primary key) |
NO |
- |
| ProcessingParentID |
int |
The ID of the superior type of the processing |
YES |
Refers to table Processing |
| DisplayText |
nvarchar (50) |
The display text of the processing as shown e.g. in a user interface |
YES |
- |
| Description |
nvarchar (MAX) |
Description of the processing |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the processing |
YES |
- |
| ProcessingURI |
varchar (255) |
A URI for a processing as defined in an external data source |
YES |
- |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entriesDefault value: (0) |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Table ProcessingMaterialCategory
The processings which are possible for a certain material category
| Column |
Data type |
Description |
Nullable |
Relation |
| ProcessingID |
int |
ID of the processing. Refers to ProcessingID in table Processing (foreign key)Default value: (1) |
NO |
Refers to table Processing |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc.Default value: N’specimen’ |
NO |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table ProjectAnalysis
The types of the analysis which are available for a project
| Column |
Data type |
Description |
Nullable |
Relation |
| AnalysisID |
int |
ID of the analysis (primary key) |
NO |
Refers to table Analysis |
| ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
NO |
Refers to table ProjectProxy |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table ProjectMaterialCategory
The material categorys which are possible for a certain Project
| Column |
Data type |
Description |
Nullable |
Relation |
| ProjectID |
int |
ID of the Project. Refers to ProjectID in table ProjectProxy (foreign key)Default value: (1) |
NO |
Refers to table ProjectProxy |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc.Default value: N’specimen’ |
NO |
Refers to table CollMaterialCategory_Enum |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollMaterialCategory_Enum
- ProjectProxy
Table ProjectProcessing
The types of processing available within a project
| Column |
Data type |
Description |
Nullable |
Relation |
| ProcessingID |
int |
ID of the table Processing (foreign key and part of primary key) |
NO |
Refers to table Processing |
| ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
NO |
Refers to table ProjectProxy |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table ProjectTaxonomicGroup
The taxonomic group which are possible for a certain Project
| Column |
Data type |
Description |
Nullable |
Relation |
| ProjectID |
int |
ID of the Project. Refers to ProjectID in table ProjectProxy (foreign key)Default value: (1) |
NO |
Refers to table ProjectProxy |
| TaxonomicGroup |
nvarchar (50) |
Taxonomic group of specimen. Examples: ‘plant’, ‘animal’, etc.Default value: N’plant’ |
NO |
Refers to table CollTaxonomicGroup_Enum |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- CollTaxonomicGroup_Enum
- ProjectProxy
Table ProjectUser
The projects which a user can access
| Column |
Data type |
Description |
Nullable |
Relation |
| LoginName |
nvarchar (50) |
A login name which the user uses to access the DivesityWorkbench, Microsoft domains, etc.. |
NO |
Refers to table UserProxy |
| ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
NO |
Refers to table ProjectProxy |
| ReadOnly |
bit |
If the user has only read access to data of this projectDefault value: (0) |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
trgInsProjectUser
Setting ReadOnly in dependence of locked projects
Table Property
The list of the properties that can be specified for the collection site
| Column |
Data type |
Description |
Nullable |
Relation |
| PropertyID |
int |
Unique ID for the property (= primary key) |
NO |
- |
| PropertyParentID |
int |
PropertyID of the superior property |
YES |
Refers to table Property |
| PropertyName |
nvarchar (100) |
Name of the system used for the description of the collection site, e.g. Chronostratigraphy |
NO |
- |
| DefaultAccuracyOfProperty |
nvarchar (50) |
The default for the accuracy of values which can be reached with this method |
YES |
- |
| DefaultMeasurementUnit |
nvarchar (50) |
The default measurement unit for the characterisation system, e.g. pH |
YES |
- |
| ParsingMethodName |
nvarchar (50) |
Internal value, specifying a programming method used for parsing text in table CollectionEventProperty |
NO |
- |
| DisplayText |
nvarchar (50) |
Short abbreviated description of the site property as displayed in the user interface |
YES |
- |
| DisplayEnabled |
bit |
Specifies, if this item is enabled to be used within the database. Properties can be disabled to avoid seeing them, but keep the definition for the future.Default value: (1) |
YES |
- |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
YES |
- |
| Description |
nvarchar (MAX) |
Description of the property |
YES |
- |
| PropertyURI |
varchar (1000) |
The URI of the property, e.g. as provided by the module DiversityScientificTerms. |
YES |
- |
| PropertyType |
nvarchar (50) |
Type of the collection site property, e.g. Chronostratigraphy |
YES |
Refers to table PropertyType_Enum |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Content
This is the default content corresponding to the default settings in the client software.
| PropertyID |
PropertyName |
Description |
ParsingMethodName |
| 1 |
European Nature Information System (EUNIS) |
European Nature Information System, http://eunis.eea.eu.int/index.jsp |
Vegetation |
| 10 |
Geographic regions |
Import aus Tabelle Naturraum der Datenbank Mykologie aus Görlitz |
Vegetation |
| 20 |
Chronostratigraphy |
Chronostratigraphie nach Angaben der Bayerischen Staatssammlung für Paläontologie |
Stratigraphy |
| 30 |
Lithostratigraphy |
Lithostratigraphie nach Angaben der Bayerischen Staatssammlung für Paläontologie |
Stratigraphy |
| 40 |
Lebensraumtypen (LfU) |
Lebensraumtypen (Bayerisches Landesamt für Umwelt) |
Vegetation |
| 50 |
Pflanzengesellschaften |
|
Vegetation |
| 60 |
Biostratigraphy |
|
Stratigraphy |
Table ReplicationPublisher
Databases providing data via replication
| Column |
Data type |
Description |
Nullable |
Relation |
| DatabaseName |
varchar (255) |
The name of the publishing database |
NO |
- |
| Server |
varchar (255) |
The name or address of the server where the publishing database is located |
NO |
- |
| Port |
smallint |
The port used by the server |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
Table Task
The Tasks of the collection
| Column |
Data type |
Description |
Nullable |
Relation |
| TaskID |
int |
ID of the Task (primary key) |
NO |
- |
| TaskParentID |
int |
The ID of the superior type of the Task |
YES |
Refers to table Task |
| DisplayText |
nvarchar (50) |
The display text of the Task as shown e.g. in a user interface |
YES |
- |
| Description |
nvarchar (MAX) |
Description of the Task |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the Task |
YES |
- |
| TaskURI |
varchar (500) |
A URI for a Task as defined in an external data source |
YES |
- |
| Type |
nvarchar (50) |
The type of the task as defined in table TaskType_EnumDefault value: ‘Task’ |
YES |
Refers to table TaskType_Enum |
| ModuleTitle |
nvarchar (50) |
The title for module related data for collection tasks. Not available in depending collection task if empty |
YES |
- |
| ModuleType |
nvarchar (50) |
The DiversityWorkbench module to which a task is related. Not available in depending collection task if empty |
YES |
Refers to table TaskModuleType_Enum |
| SpecimenPartType |
nvarchar (50) |
The description of the collection specimen part to which a task is related. Not available in depending collection task if empty |
YES |
- |
| TransactionType |
nvarchar (50) |
The description of the transaction to which a task is related. Not available in depending collection task if empty |
YES |
- |
| ResultType |
nvarchar (50) |
The display text for the results as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| DateType |
nvarchar (50) |
The date and time details defined for a task. Not available in depending collection task if empty |
YES |
Refers to table TaskDateType_Enum |
| DateBeginType |
nvarchar (50) |
The definition of the begin for date and time details defined for a task. Not available in depending collection task if empty |
YES |
- |
| DateEndType |
nvarchar (50) |
The definition of the end for date and time details defined for a task. Not available in depending collection task if empty |
YES |
- |
| NumberType |
nvarchar (50) |
The definition for the numeric value as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| BoolType |
nvarchar (50) |
The definition for the boolean value as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| MetricType |
nvarchar (50) |
The definition for the metric as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| MetricUnit |
nvarchar (50) |
The unit of the metric, e.g. °C for temperature |
YES |
- |
| DescriptionType |
nvarchar (50) |
The definition for the description as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| NotesType |
nvarchar (50) |
The definition for the notes as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| UriType |
nvarchar (50) |
The definition for the URI as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| ResponsibleType |
nvarchar (50) |
The definition for the responsible agent as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: dbo.UserID() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: dbo.UserID() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- TaskDateType_Enum
- TaskModuleType_Enum
- TaskType_Enum
Table TaskModule
Data from DiversityWorkbench modules used used within a task
| Column |
Data type |
Description |
Nullable |
Relation |
| TaskID |
int |
ID of the task, part of PK, relates to PK of table Task |
NO |
Refers to table Task |
| DisplayText |
nvarchar (400) |
Display text as provided by the module, part of PK |
NO |
- |
| URI |
varchar (500) |
URI linking the dataset of the module |
YES |
- |
| Description |
nvarchar (MAX) |
Optional description of the linked data, e.g. the common name for taxa |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes related to the dataset |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
ID of the creator of this data setDefault value: dbo.UserID() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
ID of the person to update this data set lastDefault value: dbo.UserID() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table TaskResult
List or results provided for the CollectionTask of the corresponding type
| Column |
Data type |
Description |
Nullable |
Relation |
| TaskID |
int |
ID of the task, part of PK, relates to PK of table Task |
NO |
Refers to table Task |
| Result |
nvarchar (400) |
The result, part of PK |
NO |
- |
| URI |
varchar (500) |
A URI of the result providing further information |
YES |
- |
| Description |
nvarchar (MAX) |
The description of the entry |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes about the entry |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
ID of the creator of this data setDefault value: dbo.UserID() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
ID of the person to update this data set lastDefault value: dbo.UserID() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table Transaction
Transactions like loan, borrow, gift, exchange etc. of specimen, if they are e.g. permanently or temporary transfered from one collection to another
| Column |
Data type |
Description |
Nullable |
Relation |
| TransactionID |
int |
Unique ID for the transaction (= primary key) |
NO |
- |
| ParentTransactionID |
int |
The ID of a preceeding transaction of a superior transaction, if transactions are organized in a hierarchy |
YES |
Refers to table Transaction |
| TransactionType |
nvarchar (50) |
Type of the transaction, e.g. gift in or out, exchange in or out, purchase in or outDefault value: N’exchange’ |
NO |
Refers to table CollTransactionType_Enum |
| TransactionTitle |
nvarchar (200) |
The title of the transaction as e.g. shown in an user interface |
NO |
- |
| ReportingCategory |
nvarchar (50) |
A group defined for the transaction, e.g. a taxonomic group as used for exchange balancing |
YES |
- |
| AdministratingCollectionID |
int |
ID of the collection which is responsible for the administration of the transaction. |
NO |
Refers to table Collection |
| MaterialDescription |
nvarchar (MAX) |
Description of the material of this transactionDefault value: '’ |
YES |
- |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc.Default value: N’specimen’ |
YES |
- |
| MaterialCollectors |
nvarchar (MAX) |
The collectors of the material |
YES |
- |
| MaterialSource |
nvarchar (500) |
The source of the material within a transaction, e.g. a excavation |
YES |
- |
| FromCollectionID |
int |
The ID of the collection from which the specimen were transfered, e.g. the donating collection of a gift |
YES |
Refers to table Collection |
| FromTransactionPartnerName |
nvarchar (255) |
Name of the person or institution from which the specimen were transfered, e.g. the donator of a gift |
YES |
- |
| FromTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
YES |
- |
| FromTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the source of the specimen, e.g. the donating collection of a gift |
YES |
- |
| ToCollectionID |
int |
The ID of the collection to which the specimen were transfered, e.g. the receiver of a gift |
YES |
Refers to table Collection |
| ToTransactionPartnerName |
nvarchar (255) |
Name of the person or institution to which the specimen were transfered, e.g. the receiver of a gift |
YES |
- |
| ToTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
YES |
- |
| ToTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the destination of the specimen, e.g. the receiving collection of a gift |
YES |
- |
| NumberOfUnits |
int |
The number of units which were (initially) included in the transaction |
YES |
- |
| Investigator |
nvarchar (200) |
The investigator for whose study a transacted material was sent |
YES |
- |
| TransactionComment |
nvarchar (MAX) |
Comments on the exchanged material addressed to the transaction partner |
YES |
- |
| BeginDate |
datetime |
Date when the transaction started |
YES |
- |
| AgreedEndDate |
datetime |
End of the transaction period, e.g. if the time for borrowing the specimen is restricted |
YES |
- |
| ActualEndDate |
datetime |
Actual end of the transaction after a prolonation when e.g. the date of return for a loan was prolonged by the owner |
YES |
- |
| DateSupplement |
nvarchar (100) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. |
YES |
- |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction not to be published e.g. on a web page |
YES |
- |
| ToRecipient |
nvarchar (255) |
The recipient receiving the transaction e.g. if not derived from the link to DiversityAgents |
YES |
- |
| ResponsibleName |
nvarchar (255) |
The person responsible for this transaction |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
The URI of the person, team or organisation responsible for the data (see e.g. module DiversityAgents) |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: user_name() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: user_name() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
- Collection
- CollTransactionType_Enum
Table TransactionAgent
Agents involved in the transaction
| Column |
Data type |
Description |
Nullable |
Relation |
| TransactionID |
int |
Unique ID for the transaction, refers to table Transaction (= part of primary key and foreign key) |
NO |
Refers to table Transaction |
| TransactionAgentID |
int |
Unique ID for the Agent within the transaction (= part of primary key) |
NO |
- |
| AgentName |
nvarchar (500) |
Name of the person or institution |
YES |
- |
| AgentURI |
varchar (500) |
Link to the source for further informations about the agent, e.g in the module DiversityAgents |
YES |
- |
| AgentRole |
nvarchar (500) |
Role of the agent within the transaction |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes about the agent |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
The standard text phrases for transactions
| Column |
Data type |
Description |
Nullable |
Relation |
| Comment |
nvarchar (400) |
Text as transferred into the comment of a transaction |
NO |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Table TransactionDocument
The history of transactions or the documents connected to the transactions
| Column |
Data type |
Description |
Nullable |
Relation |
| TransactionID |
int |
Unique ID for the transaction, refers to table Transaction (= part of primary key and foreign key) |
NO |
Refers to table Transaction |
| Date |
datetime |
The date of the event of a transaction |
NO |
- |
| TransactionText |
nvarchar (MAX) |
The text of a transaction document |
YES |
- |
| TransactionDocument |
image |
A scanned document connected to this transaction |
YES |
- |
| DisplayText |
nvarchar (255) |
A display text as shown e.g. in a user interface to characterize the document |
YES |
- |
| DocumentURI |
varchar (1000) |
A link to a web resource of the document the document |
YES |
- |
| DocumentType |
nvarchar (255) |
The type of the document |
YES |
- |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Table TransactionPayment
The payments within a transaction
| Column |
Data type |
Description |
Nullable |
Relation |
| TransactionID |
int |
Unique ID for the transaction, refers to table Transaction (= part of primary key and foreign key) |
NO |
Refers to table Transaction |
| PaymentID |
int |
Unique ID for the payment (= part of primary key) |
NO |
- |
| Identifier |
nvarchar (500) |
An identifer for the payment like a booking number or invoice number |
YES |
- |
| Amount |
float |
Amount of the payment in the default currency as defined in TransactionCurrency |
YES |
- |
| ForeignAmount |
float |
If the payment was not in the default curreny as defined in TransactionCurrency, the amount of the payment in foreign curreny |
YES |
- |
| ForeignCurrency |
nvarchar (50) |
If the payment was not in the default curreny as defined in TransactionCurrency, the foreign currency of the payment |
YES |
- |
| PayerName |
nvarchar (500) |
Name of the person or institution paying the amount |
YES |
- |
| PayerAgentURI |
varchar (500) |
Link to the source for further infomations about the payer, e.g in the module DiversityAgents |
YES |
- |
| RecipientName |
nvarchar (500) |
Agent receiving the payment |
YES |
- |
| RecipientAgentURI |
varchar (500) |
Link to the source for further infomations about the recipient of the payment, e.g in the module DiversityAgents |
YES |
- |
| PaymentDate |
datetime |
Date of the payment |
YES |
- |
| PaymentDateSupplement |
nvarchar (50) |
Supplement to the date of the payment, e.g. if the original date is not a real date like ‘summer 1920’ or ‘1910 - 1912’ |
YES |
- |
| PaymentURI |
varchar (500) |
A link to an external administration system for the payment |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes about the payment |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data setDefault value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated lastDefault value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set lastDefault value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
NO |
- |
Depending on:
Diversity Collection
Enumeration tables
The following objects are not included:
- Logging tables
- System objects
- Objects marked as obsolete
- Previous versions of objects
Table
| Column |
Data type |
Description |
| Code |
nvarchar (50) |
A text code that uniquely identifies each object in the enumeration (primary key). This value may not be changed, because the application may depend upon it. |
| ParentCode |
nvarchar (50) |
The code of the superior entry, if a hierarchy within the entries is necessary |
| ParentRelation |
nvarchar (50) |
Relation to parent entry, e.g. part of |
| Description |
nvarchar (500) |
Description of enumerated object, displayed in the user interface |
| DisplayText |
nvarchar (50) |
Short abbreviated description of the object, displayed in the user interface |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
| DisplayEnable |
bit |
Enumerated objects can be hidden from the user interface if this attribute is set to false (= unchecked check box) |
| URL |
varchar (500) |
A link to further information about the enumerated object |
| InternalNotes |
nvarchar (500) |
Internal development notes about usage, definition, etc. of an enumerated object |
| Icon |
image |
A symbol representing this entry in the user interface |
| ModuleName |
varchar (50) |
If the enumerated entry is related to a DiversityWorkbench module or a related webservice, the name of the DiversityWorkbench module, e.g. DiversityCollEventSeriess |
| RowGUID |
uniqueidentifier |
-Default value: newsequentialid() |
Table AnnotationType_Enum
The types of an annotation as used in table Annotation
Dependent tables:
Table CollCircumstances_Enum
Circumstances of the occurence of the organisms
Dependent tables:
Content
| Code |
Description |
| bred |
animal born in captivity (domestic, zoo, or farm animales) |
| captivity |
wild animal or plant raised in captivity |
| cultivated |
cultivated in enclosed conditions (greenhouse plants, fungi, bacteria, etc. in laboratory or biotechnology conditions) |
| experiment |
material cultivated or treated under experimental conditions (including, e.g., infection experiments) |
| imported |
imported material of uncertain provenence (e.g. food items) |
| natural |
occurrence under natural conditions |
| planted |
planted in open agriculture, horticulture, or silviculture |
| unknown |
unknown circumstances of occurrence |
Table CollCollectionImageType_Enum
The types of a collection, e.g. cupboard, drawer, box, rack etc.
Dependent tables:
Content
| Code |
Description |
| area |
area resp. section within a collection, e.g. within a insect case |
| image |
image of a collection, e.g. a insect case |
| plan |
Old plan optional including geometry of a collection |
Table CollCollectionType_Enum
The types of a collection, e.g. cupboard, drawer, box, rack etc.
Dependent tables:
| Code |
Description |
DisplayText |
| area |
An area within a collection, e.g. within an insect display case |
area |
| box |
box |
box |
| collection |
A collection of objects |
collection |
| container |
container |
container |
| cupboard |
cupboard |
cupboard |
| department |
department |
department |
| drawer |
drawer |
drawer |
| freezer |
freezer |
freezer |
| fridge |
fridge |
fridge |
| hardware |
hardware |
hardware e.g. for sensor network |
| institution |
institution |
institution |
| location |
location |
location |
| radioactive |
Place where radioactive objects are stored |
radioactive |
| room |
room |
room |
| sensor |
A sensor for e.g. temperature or pests |
sensor |
| steel locker |
steel locker |
steel locker |
| subdivided container |
A container subdivided into chambers |
subdivided container |
| trap |
A trap for monitoring pest e.g. for IPM |
trap |
Table CollDateCategory_Enum
The categories for the collection date
Dependent tables:
Table CollEventDateCategory_Enum
The categoies of the collection date
Dependent tables:
Content
| Code |
Description |
| actual |
The given date is the real date |
| estimated |
The given date was estimated by the user |
| system |
The given date was generated by the system |
Table CollEventImageType_Enum
The types of the images taken from a collection event
Dependent tables:
Content
| Code |
Description |
DisplayText |
ParentCode |
| aerial photography |
photography from an elevated position e.g. an aerial drone |
aerial photography |
|
| audio |
audio file related to an event |
sound |
|
| audio (for transcription) |
audio file of spoken comments related to an event that might be deleted after transcription into the database |
sound /recorded speech |
audio |
| biotope photography |
photography of a whole biotope where the specimen are observed resp. collected |
biotope photography |
|
| documentation |
a still image of a document or paperwork associated with an event, e. g. field books |
documentation |
|
| drawing, painting |
original line or color drawing of an event |
drawing, painting |
|
| image |
still image of any raster or vector format without any further information regarding the content type, e. g. graphic designs, plans and maps, vector graphics 2D, 3D models |
image |
|
| landscape photography |
photography of the landscape where the specimen are observed resp. collected |
landscape photography |
|
| map |
image of a map where the specimen are observed resp. collected |
map |
image |
| photograph |
photographic still image of an event; e. g. trap, trawl net |
photograph |
|
| supporting files |
text-xml or text-WKT encoded files, assigned to multimedia objects |
supporting files |
|
| video |
e. g. video file related to an event |
moving image |
|
Table CollEventSeriesDescriptorType_Enum
The type of the Descriptors
Dependent tables:
- CollectionEventSeriesDescriptor
Table CollEventSeriesImageType_Enum
The types of the images taken from a collection event series
Dependent tables:
- CollectionEventSeriesImage
Content
| Code |
Description |
DisplayText |
ParentCode |
| aerial photography |
photography from an elevated position e.g. an aerial drone |
aerial photography |
|
| audio |
audio record of e.g. the song of an observed bird |
sound |
|
| audio (for transcription) |
audio file of spoken comments related to an event or event series that might be deleted after transcription into the database |
sound /recorded speech |
audio |
| documentation |
a still image of a document or paperwork associated with an event series, e. g. field books |
documentation |
|
| drawing, painting |
original line or color drawing of an event series |
drawing, painting |
|
| image |
still image of any raster or vector format without any further information regarding the content type, e. g. graphic designs, plans and maps, vector graphics 2D, 3D models |
image |
|
| landscape photography |
photography of the landscape where the specimen are observed resp. collected |
landscape photography |
|
| map |
image of a map where the specimen are observed resp. collected |
map |
image |
| moving image |
e. g. video file related to an event series |
moving image |
|
| photograph |
photographic still image of an event series; e. g. trap, trawl net |
photograph |
|
| supporting files |
text-xml or text-WKT encoded files, assigned to multimedia objects |
supporting files |
|
Table CollIdentificationCategory_Enum
The categories of an identification, e.g. determination, confirmation etc.
Dependent tables:
Content
| Code |
Description |
| absence |
When nomenclatural type material has been used up in previous studies, or if a mixed collection contains e.g. several fungi but none fits the protologue |
| confirmation |
The examination confirms a previous identification |
| correction |
Correction of a misidentification supposedly due to a typing error. |
| determination |
The determination of the material results in a new identification or correction |
| dubious |
The material is assumed as dubiously belonging to the identified taxon or the observation is assumed as dubiously belonging to the identified taxon |
| expert assignment |
The assignment of occurrence data to a new taxon following expert opinion, without study of material |
| implicit |
The identification can be logically deduced from the type material, e.g. in the case of Yes/No type specimens. |
| negative |
The material is identified as not belonging to the identified taxon or the observation is verified as not belonging to the identified taxon |
| preference |
Assigning preference to an older determination over the most recent one. Differs from a confirmation only in that the object has not been re-examined. The choice is based on the expertise of the person effecting the earlier identification |
| renaming |
e.g. assigning a species to a new genus or a purely nomenclatural change |
| revision |
The correction of a former determination of the material |
| type annotation |
annotated as original material (e. g. syntype, paratype), e.g. done by a scientist by adding type labels to the material as part of a curatorial process |
| type designation |
designated as type material (i. e. lectotype, neotype, epitype or their duplicates; ICBN 2006, Art. 7.5, 7.10-11 + N2, Art. 9.7-11, 9.20, 9A, Art. 37.3; nomenclatural action undertaken by the person who is treating the nomenclature of the taxon) |
| type indication |
indicated as type material (ICBN 2006, Art. 37.1-4, 37A, Art. 7.5, Art. 24.3; any nomenclatural action, e. g. by the person who described the taxon, other than type designation) |
Table CollIdentificationDateCategory_Enum
The categories of the identification date
Dependent tables:
| Code |
Description |
| actual |
The given date is the real date |
| collection date |
The collection date has been taken as the identification date |
| estimated |
The given date was estimated by the user |
| system |
The given date was generated by the system |
Table CollIdentificationQualifier_Enum
The identification qualifiers, e.g. cf. spec., s.l. etc.
Dependent tables:
Content
| Code |
Description |
| (select this for identifications that are certain) |
|
| ? |
unspecified doubtful identification (aff. or cf., or applicable taxonomic rank not known) |
| aff. forma |
‘affinis’ = similar to, but probably not the named forma |
| aff. gen. |
‘affinis’ = similar to, but probably not the named genus |
| aff. sp. |
‘affinis’ = similar to, but probably not the named species |
| aff. subsp. |
‘affinis’ = similar to, but probably not the named subspecies |
| aff. var. |
‘affinis’ = similar to, but probably not the named variety |
| agg. |
aggregate |
| cf. forma |
doubtful identification of forma or forma specialis. Example: ‘Ech. ferruginea cf. f. rubra’ |
| cf. gen. |
doubtful genus identification. Examples: ‘cf. Abies’, ‘?Abies’ |
| cf. hybrid |
doubtful identification of hybrid. Example: cf. Salix alba x fragilis |
| cf. sp. |
doubtful species identification (genus identification certain). Example: ‘Abies cf. alba’ |
| cf. subsp. |
doubtful identification of subspecies. Example: ‘Abies alba cf. ssp. alba’ |
| cf. var. |
doubtful identification of variety. Example: ‘Echinonema ferruginea cf. var. campestris’ |
| s. l. |
sensu lato |
| s. str. |
sensu stricto |
| sp. |
only the genus is given, the information relates to a single, unspecified species |
| sp. nov. |
only the genus is given, it is assumed that the unidentified species is new |
| spp. |
only the genus is given, the information covers several unspecified species |
Table CollLabelTranscriptionState_Enum
The states of the transcription of a label
Dependent tables:
Content
| Code |
Description |
| complete |
all data from label transferred in database |
| curator review required |
curator for transcription / review required |
| extern review required |
extern curator for transcription / review required |
| final curator review |
specimen has been finally reviewed by curator |
| first curator review |
specimen has been reviewed by curator for first time |
| incomplete |
transcription started by technical staff |
| not started |
transcription not started |
Table CollLabelType_Enum
The types of a label, e.g. handwritten, typed etc.
Dependent tables:
Content
| Code |
Description |
DisplayText |
| handwriting |
all data in handwriting |
handwriting |
| mixed |
data typed with handwriting comments |
mixed |
| typed |
all data typed |
typed |
Table CollMaterialCategory_Enum
The material categories of the specimen, e.g. slide, culture etc.
Dependent tables:
- CollectionSpecimenPart
- ProjectMaterialCategory
Content
| Code |
Description |
DisplayText |
ParentCode |
| bones |
bones or skeleton from vertebrates |
bones |
preserved specimen |
| complete skeleton |
complete skeleton |
complete skeleton |
bones |
| cones |
cones of e.g. Gymnospermae, not stored together with the herbarium sheet |
cones |
preserved specimen |
| cultures |
living cultures of organisms |
cultures |
living specimen |
| DNA lyophilised |
DNA lyophilised |
DNA lyophilised |
DNA sample |
| DNA sample |
DNA sample |
DNA sample |
other specimen |
| drawing |
original line or color drawing |
drawing |
drawing or photograph |
| drawing or photograph |
A record describing a static visual representation (digital or physical) |
drawing or photograph |
medium |
| dried specimen |
dried specimen |
dried specimen |
preserved specimen |
| earth science specimen |
A record describing a geo-scientific object which is no pure mineral, rock or fossil specimen (but a mixture), or a geo-scientific specimen with unknown traits |
earth science specimen |
other specimen |
| egg |
egg, e.g. of a bird or insect |
egg |
other specimen |
| fossil bones |
fossil bones or skeleton from vertebrates |
fossil bones |
fossil specimen |
| fossil complete skeleton |
fossil complete skeleton of a vertrebrate |
fossil complete skeleton |
fossil bones |
| fossil incomplete skeleton |
fossil incomplete skeleton of a vertrebrate |
fossil incomplete skeleton |
fossil bones |
| fossil otolith |
fossil otolith of fishes |
fossil otolith |
fossil bones |
| fossil postcranial skeleton |
fossil postcranial skeleton of a vertrebrate |
fossil postcranial skeleton |
fossil bones |
| fossil scales |
fossil scales of fish |
fossil scales |
fossil specimen |
| fossil shell |
fossil shell of animal |
fossil shell |
fossil specimen |
| fossil single bones |
fossil single bones of a vertrebrate |
fossil single bones |
fossil bones |
| fossil skull |
fossil skull of a vertrebrate |
fossil skull |
fossil bones |
| fossil specimen |
A record describing a preserved specimen that is a fossil |
fossil specimen |
specimen |
| fossil tooth |
fossil tooth of an animal |
fossil tooth |
fossil bones |
| herbarium sheets |
capsules or sheets as stored in a botanical collection |
herbarium sheets |
dried specimen |
| human observation |
A record describing an output of a human observation process |
human observation |
observation |
| icones |
icones, images etc. stored in a biological collection |
icones |
drawing or photograph |
| incomplete skeleton |
incomplete skeleton |
incomplete skeleton |
bones |
| living specimen |
A record describing a specimen which is alive (not preserved) |
living specimen |
specimen |
| machine observation |
A record describing an output of a machine observation process |
machine observation |
observation |
| material sample |
A record describing the physical result of a sampling (or subsampling) event (sample either preserved or destructively processed) |
material sample |
other specimen |
| medium |
medium like sound recordings, videos, images and the like |
medium |
|
| micr. slide |
glass plate with specimen for microscopic study |
micr. slide |
other specimen |
| mineral specimen |
A record describing a preserved specimen which is a mineral |
mineral specimen |
other specimen |
| mould |
mould of animal |
mould |
fossil specimen |
| nest |
nest, e.g. of a bird or insect |
nest |
other specimen |
| observation |
observation |
observation |
|
| other specimen |
A record describing a specimen which is not indicated as preserved, living, fossil or mineral |
other specimen |
specimen |
| otolith |
otolith of fishes |
otolith |
bones |
| pelt |
pelt, skin or fur of an animal |
pelt |
preserved specimen |
| photogr. print |
photographic print (color or black/white) |
photogr. print |
drawing or photograph |
| photogr. slide |
image in the form of a photographic slide for projection |
photogr. slide |
drawing or photograph |
| pinned specimen |
pinned specimen, e.g. insects in a insect case |
pinned specimen |
dried specimen |
| postcranial skeleton |
postcranial skeleton |
postcranial skeleton |
bones |
| preserved specimen |
A record describing a preserved specimen (not living) |
preserved specimen |
specimen |
| SEM table |
table with specimen for Scanning electron microscopy |
SEM table |
other specimen |
| shell |
shell e.g. of a snail |
shell |
preserved specimen |
| single bones |
single bones |
single bones |
bones |
| skull |
skull of a vertebrate |
skull |
bones |
| sound |
sound recoding |
sound |
medium |
| specimen |
specimen stored in a collection |
specimen |
|
| TEM specimen |
specimen preparation for Transmission electron microscopy |
TEM specimen |
other specimen |
| thin section |
thin section |
thin section |
other specimen |
| tissue sample |
tissue sample |
tissue sample |
other specimen |
| tooth |
tooth of an animal |
tooth |
bones |
| trace |
trace e.g. footprints of an animal |
trace |
other specimen |
| trace fossil |
trace fossil of animal |
trace fossil |
fossil specimen |
| vial |
vial, flask or similar container |
vial |
preserved specimen |
Table CollRetrievalType_Enum
The types data about organisms were retrieved
Dependent tables:
Content
| Code |
Description |
| human observation |
A record describing an output of a human observation process |
| literature |
A record describing an output of a literature research |
| machine observation |
A record describing an output of a machine observation process |
Table CollSpecimenImageType_Enum
The type of an image of a specimen, e.g. label
Dependent tables:
Content
| Code |
Description |
DisplayText |
ParentCode |
| audio |
e. g. audio file related to a specimen, observation or unit |
sound |
|
| audio (for transcription) |
e. g. audio file of spoken comments related to a specimen, observation or unit that might be deleted after transcription into the database |
sound /recorded speech |
audio |
| drawing |
original line or color drawing and painting of a specimen, observation or unit |
drawing, painting |
image |
| image |
still image of any raster or vector format without any further information regarding the content type, e. g. graphic designs, plans and maps, vector graphics 2D, 3D models |
image |
|
| label |
a still image of a document or paperwork associated with a specimen, observation or unit, e. g. herbarium label, file card |
documentation |
image |
| LM photograph |
photographic still image of a specimen, observation or unit from light microscopy |
LM photograph |
image |
| photograph |
photographic still image of a specimen, observation or unit; e. g. macro images of an object |
photograph |
|
| SEM image |
photographic still image of a specimen, observation or unit from scanning electron microscopy |
SEM photograph |
image |
| supporting files |
text-xml or text-WKT encoded files, assigned to multimedia objects |
supporting files |
|
| TEM image |
photographic still image of a specimen, observation or unit from transmission electron microscopy |
TEM photograph |
image |
| video |
e. g. video file related to an specimen, observation or unit |
moving image |
|
Table CollSpecimenRelationType_Enum
Types of the relation between specimen
Dependent tables:
- CollectionSpecimenRelation
Content
| Code |
Description |
| Duplicate |
specimen or specimen part from the same location and time, designated by the same collector as such (may be one organism or not) |
| public database |
public database |
| Same location |
different specimens from the same location, but potentially different time and collector |
| Specimen part |
material originated from one specimen, e.g. dried material + cultured material or wet material + DNA or tissue sample |
Table CollTaskMetricAggregation_Enum
The aggregation applied for retrieval of values, e.g. avg, max etc.
Dependent tables:
Content
| Code |
Description |
| avg |
the average value of a period |
| max |
the maximal value within a period |
| min |
the minimal value within a period |
| none |
no aggregation applied |
| range |
the difference between the maximal and minimal value within a period |
| sum |
the sum of values within a period |
Table CollTaxonomicGroup_Enum
The taxonomic groups of the organisms
Dependent tables:
- IdentificationUnit
- ProjectTaxonomicGroup
Content
| Code |
Description |
DisplayText |
ParentCode |
| alga |
alga |
alga |
|
| amphibian |
amphibian |
amphibian |
vertebrate |
| animal |
Animalia (= Metazoa) |
animal |
|
| artefact |
An item made or given shape by humans, such as a tool or a work of art |
artefact |
|
| arthropod |
Arthropoda: insects, spiders, crabs etc. |
arthropod |
evertebrate |
| bacterium |
bacteria/prokaryotic |
bacterium |
|
| bird |
bird |
bird |
vertebrate |
| bryophyte |
Marchantiomorpha (liverworts), Anthocerotophyta (hornworts) and Bryophyta (mosses) |
bryophyte |
|
| cnidaria |
Cnidaria: sea anemones, corals, jellyfish, sea pens, hydra |
cnidaria |
evertebrate |
| Coleoptera |
Coleoptera |
Coleoptera |
insect |
| Diptera |
Diptera |
Diptera |
insect |
| echinoderm |
Echinodermata: sea urchins, starfish, and their allies |
echinoderm |
evertebrate |
| evertebrate |
Animalia (= Metazoa) excl. Chordata |
evertebrate |
animal |
| fish |
fish |
fish |
vertebrate |
| fungus |
Eumycota: mushrooms, sac fungi, yeast, molds, rusts, smuts, etc. |
fungus |
|
| gall |
growth modification or deformation of plant tissue or fungal plectenchyma, induced by a different organism |
gall |
|
| Heteroptera |
Heteroptera |
Heteroptera |
insect |
| Hymenoptera |
Hymenoptera |
Hymenoptera |
insect |
| insect |
Hexapoda: insects, springtails, diplurans, and proturans |
insect |
arthropod |
| Lepidoptera |
Lepidoptera |
Lepidoptera |
insect |
| lichen |
Lichenized Eumycota |
lichen |
|
| mammal |
mammal |
mammal |
vertebrate |
| mineral |
A record describing a preserved specimen which is a mineral |
mineral |
|
| mollusc |
Mollusca: Snails, clams, mussels, squids, octopi, chitons, and tusk shells |
mollusc |
evertebrate |
| myxomycete |
Myxomycota |
slime mould |
|
| other |
other/anorganic |
other |
|
| plant |
Lycophytes, ferns and seedplants |
plant |
|
| reptile |
reptile |
reptile |
vertebrate |
| rock |
A record describing a preserved specimen which is a rock |
rock |
|
| soil |
soil |
soil |
|
| soil horizon |
soil horizon |
soil horizon |
soil |
| spider |
Spider: Including Araneae (Mesothelae, Mygalomorphae and Araneomorphae) |
spider |
arthropod |
| unknown |
unknown group |
unknown |
|
| vertebrate |
Chordata |
vertebrate |
animal |
| virus |
virus |
virus |
|
Table CollTransactionType_Enum
The types of the transactions of the specimen, e.g. loan, exchange
Dependent tables:
Content
| Code |
Description |
| embargo |
Temporary data embargo for specimen that should not be published within the specified period |
| exchange |
Exchange of specimens between e.g. collections |
| forwarding |
a forwarding of specimens of a loan to another institution |
| gift |
A gift of specimens to e.g. a collection |
| inventory |
An inventory of e.g. a collection |
| loan |
A temporary loan of specimens of a collection to e.g. another collection |
| permanent loan |
A permanent loan of specimens of a collection to e.g. another collection |
| permit |
permit or certificate for the collection of specimen |
| purchase |
A purchase of specimens by a collection |
| regulation |
a regulation, provisio or stipulation e.g. in the context of the Nagoya protocoll |
| removal |
the removal of specimens from an institution, e.g. by damage, loss etc. |
| return |
a partial return of a loan |
| transaction group |
the superior dataset for a group of transactions |
| warning |
A warning e.g. against biocides in samples |
Table CollTypeStatus_Enum
The type statuses of the identifications, e.g. Isotype
Dependent tables:
Content
| Code |
Description |
| allolectotype |
A paralectotype specimen that is the opposite sex of the lectotype. The term is not regulated by the ICZN. [Zoo.] |
| alloneotype |
A paraneotype specimen that is the opposite sex of the neotype. The term is not regulated by the ICZN. [Zoo.] |
| allotype |
A paratype specimen designated from the type series by the original author that is the opposite sex of the holotype. The term is not regulated by the ICZN. [Zoo.] |
| conserved type |
The one specimen or other element conserved as type for a name of a subdivision of a genus or of an infraspecific taxonand listed in ICN, App. III and IV (typ. cons.) |
| cotype |
A deprecated term no longer recognized in the ICZN; formerly used for either syntype or paratype [see ICZN Recommendation 73E]. [Zoo.] |
| epitype |
An epitype is a specimen or illustration selected to serve as an interpretative type when any kind of holotype, lectotype, etc. is demonstrably ambiguous and cannot be critically identified for purposes of the precise application of the name of a taxon (see Art. ICBN 9.7, 9.18). An epitype supplements, rather than replaces existing types. [Bot./Bio.] |
| ex-Epitype |
A strain or cultivation derived from epitype material. Ex-types are not regulated by the botanical or zoological code. [Bot.] |
| ex-Holotype |
A strain or cultivation derived from holotype material. Ex-types are not regulated by the botanical or zoological code. [Zoo./Bot.] |
| ex-Isotype |
A strain or cultivation derived from isotype material. Ex-types are not regulated by the botanical or zoological code. [Zoo./Bot.] |
| ex-Lectotype |
A strain or cultivation derived from lectotype material. Ex-types are not regulated by the botanical or zoological code. [Zoo./Bot.] |
| ex-Neotype |
A strain or cultivation derived from neotype material. Ex-types are not regulated by the botanical or zoological code. [Zoo./Bot.] |
| ex-Paratype |
A strain or cultivation derived from paratype material. Ex-types are not regulated by the botanical or zoological code. [Zoo./Bot.] |
| ex-Syntype |
A strain or cultivation derived from neotype material. Ex-types are not regulated by the botanical or zoological code. [Zoo./Bot.] |
| ex-Type |
A strain or cultivation derived from some kind of type material. Ex-types are not regulated by the botanical or zoological code. [Zoo./Bot.] |
| hapantotype |
One or more preparations of directly related individuals representing distinct stages in the life cycle, which together form the type in an extant species of protistan [ICZN Article 72.5.4]. A hapantotype, while a series of individuals, is a holotype that must not be restricted by lectotype selection. If a hapantotype is found to contain individuals of more than one species, however, components may be excluded until it contains individuals of only one species [ICZN Article 73.3.2]. [Zoo.] |
| holotype |
The one specimen or other element used or designated by the original author at the time of publication of the original description as the nomenclatural type of a species or infraspecific taxon. A holotype may be ’explicit’ if it is clearly stated in the originating publication or ‘implicit’ if it is the single specimen proved to have been in the hands of the originating author when the description was published. [Zoo./Bot./Bio.] |
| iconotype |
A drawing or photograph (also called ‘phototype’) of a type specimen. Note: the term “iconotype” is not used in the ICBN, but implicit in, e. g., ICBN Art. 7 and 38. [Zoo./Bot.] |
| isoepitype |
a duplicate of an epitype |
| isolectotype |
A duplicate of a lectotype, compare lectotype. [Bot.] |
| isoneotype |
A duplicate of a neotype, compare neotype. [Bot.] |
| isoparatype |
A duplicate of a paratype |
| isosyntype |
A duplicate of a syntype, compare isotype = duplicate of holotype. [Bot.] |
| isotype |
An isotype is any duplicate of the holotype (i. e. part of a single gathering made by a collector at one time, from which the holotype was derived); it is always a specimen (ICBN Art. 7). [Bot.] |
| lectotype |
A specimen or other element designated subsequent to the publication of the original description from the original material (syntypes or paratypes) to serve as nomenclatural type. Lectotype designation can occur only where no holotype was designated at the time of publication or if it is missing (ICBN Art. 7, ICZN Art. 74). [Zoo./Bot.] – Note: the BioCode defines lectotype as selection from holotype material in cases where the holotype material contains more than one taxon [Bio.]. |
| neotype |
A specimen designated as nomenclatural type subsequent to the publication of the original description in cases where the original holotype, lectotype, all paratypes and syntypes are lost or destroyed, or suppressed by the (botanical or zoological) commission on nomenclature. In zoology also called “Standard specimen” or “Representative specimen”. [Zoo./Bot./Bio.] |
| not a type |
For specimens erroneously labelled as types an explicit negative statement may be desirable. [General] |
| original material |
Specimens and illustrations indicated in the protologue of a name (see ICBN Art. 9 Note 2 for details) |
| paralectotype |
All of the specimens in the syntype series of a species or infraspecific taxon other than the lectotype itself. Also called “lectoparatype”. [Zoo.] |
| paraneotype |
All of the specimens in the syntype series of a species or infraspecific taxon other than the neotype itself. Also called “neoparatype”. [Zoo.] |
| paratype |
All of the specimens in the type series of a species or infraspecific taxon other than the holotype (and, in botany, isotypes). Paratypes must have been at the disposition of the author at the time when the original description was created and must have been designated and indicated in the publication. Judgment must be exercised on paratype status, for only rarely are specimens explicitly cited as paratypes, but usually as “specimens examined,” “other material seen”, etc. [Zoo./Bot.] |
| plastoholotype |
A copy or cast of holotype material (compare Plastotype). |
| plastoisotype |
A copy or cast of isotype material (compare Plastotype). |
| plastolectotype |
A copy or cast of lectotype material (compare Plastotype). |
| plastoneotype |
A copy or cast of neotype material (compare Plastotype). |
| plastoparatype |
A copy or cast of paratype material (compare Plastotype). |
| plastosyntype |
A copy or cast of syntype material (compare Plastotype). |
| plastotype |
A copy or cast of type material, esp. relevant for fossil types. Not regulated by the botanical or zoological code (?). [Zoo./Bot.] |
| secondary type |
A referred, described, measured or figured specimen in the original publication (including a neo/lectotypification publication) that is not a primary type. |
| supplementary type |
A referred, described, measured or figured specimen in a revision of a previously described taxon. |
| syntype |
One of the series of specimens used to describe a species or infraspecific taxon when neither a single holotype by the original author, nor a lectotype in a subsequent publication has been designated. The syntypes collectively constitute the name-bearing type. [Zoo./Bot.] |
| topotype |
One or more specimens collected at the same location as the type series (type locality), regardless of whether they are part of the type series. Topotypes are not regulated by the botanical or zoological code. Also called “locotype”. [Zoo./Bot.] |
| type |
a) A specimen designated or indicated any kind of type of a species or infraspecific taxon. If possible more specific type terms (holotype, syntype, etc.) should be applied. b) the type name of a name of higher rank for taxa above the species rank. [General] |
| typotype |
the specimen used to prepare an illustration where the latter is the type |
Table CollUnitRelationType_Enum
The type of relation between organisms within one specimen, e.g. growing on
Dependent tables:
Content
| Code |
Description |
| Association |
in association with (unspecific statement if no further information about interaction b/w 2 organisms is available) |
| Child of |
child association (i.e. in the literal genetical sense, not in an abstract sense) |
| Endophytic in |
endophytic organism growing in plants without causing symptoms (possibly mutualistic or opportunistic parasite) |
| Found on |
Organism found on 2nd organism |
| Gall inducing |
Growth movement or swelling growth of (pseudo-)tissue or organ of a host organism caused by an organism. |
| Growing on |
substrate statement: organism growing on 2nd organism, living stages directly observed (i.e. assoc. not due to diaspores alone) |
| Isolated from |
substrate statement: organism isolated from 2nd organism, no living stages observed (i.e. possibly due to surface contamination) |
| Lichenization |
mutualistic symbiosis between a fungus and algae (= 2nd organism) |
| Mutant of |
A mutant of the parent organism |
| Mutualism |
mutualistic symbiosis, both partners profit from each other |
| Mycorrhiza of |
mycorrhizal association of a fungus with a plant (= 2nd organism) |
| Parasitic (accid.) |
parasitic on an ‘accidential host’ (in the sense of Nannfeldt): with strong infection pressure, another organism growing between infected organisms may become infected. |
| Parasitic (facul.) |
facultatively parasitic on host (= 2nd organism) |
| Parasitic (oblig.) |
obligately parasitic on host (= 2nd organism) |
| Parasitic (quest.) |
questionably parasitic on host (= 2nd organism), parasitic interaction is presumed, but doubtful/unverified |
| Parasitic on |
the taxon is parasitizing its host (regardless of type, i.e. holo-/meta-/hemi-biotrophic, or pertho-/necrotrophic) |
| Parent/Child |
parent/child association (i.e. in the literal genetical sense, not in an abstract sense) |
| Part of |
belongs to the same organism resp. unit, e.g. a branch is part of a tree |
| Pollinator of |
organism 1 is the pollinator of a plant (organism 2) |
| Predator/Food |
organism 1 is the predator of organism 2 (i. e. 2 is food of 1) |
| Saprophytic on |
organism growing saprophytic on 2nd organism, living stages directly observed (i.e. assoc. not due to diaspores alone) |
| Sibling |
both organisms are siblings (have the same parents) |
Table EntityAccessibility_Enum
The type of accessibility of database objects within a certain context
Dependent tables:
Table EntityContext_Enum
The context in which the application is used, e.g. collection - administration, field-mapping
Dependent tables:
- EntityRepresentation
- EntityUsage
Table EntityDetermination_Enum
The way a value of an entry is determined, e.g. calculated, defined by the user etc.
Dependent tables:
Table EntityLanguageCode_Enum
The codes for the languages used for the entities
Dependent tables:
Table EntityUsage_Enum
The usage of an entity, e.g. hidden, readonly
Table EntityVisibility_Enum
Type of visibility of an entity as used in table EntityUsage
Dependent tables:
Table LanguageCode_Enum
The codes for the languages
Content
| Code |
DisplayText |
| af |
Afrikaans |
| ar |
Arabisch |
| az |
Aserbaidschanisch |
| be |
Belarussisch |
| bg |
Bulgarisch |
| ca |
Katalanisch |
| cs |
Tschechisch |
| da |
Dänisch |
| de |
Deutsch |
| el |
Griechisch |
| en |
Englisch |
| es |
Spanisch |
| et |
Estnisch |
| eu |
Baskisch |
| fa |
Farsi |
| fi |
Finnisch |
| fo |
Färingisch |
| fr |
Französisch |
| gl |
Galizisch |
| gu |
Gujarati |
| he |
Hebräisch |
| hi |
Hindi |
| hr |
Kroatisch |
| hu |
Ungarisch |
| hy |
Armenisch |
| id |
Indonesisch |
| is |
Isländisch |
| it |
Italienisch |
| ja |
Japanisch |
| ka |
Georgisch |
| kk |
Kasachisch |
| kn |
Kannada |
| ko |
Koreanisch |
| ky |
Kirgisisch |
| lt |
Litauisch |
| lv |
Lettisch |
| mk |
Mazedonisch |
| mn |
Mongolisch |
| mr |
Marathi |
| ms |
Malaiisch |
| nl |
Niederländisch |
| no |
Norwegisch |
| pa |
Punjabi |
| pl |
Polnisch |
| pt |
Portugiesisch |
| ro |
Rumänisch |
| ru |
Russisch |
| sa |
Sanskrit |
| sk |
Slowakisch |
| sl |
Slowenisch |
| sq |
Albanisch |
| sv |
Schwedisch |
| sw |
Swahili |
| ta |
Tamil |
| te |
Telugu |
| th |
Thai |
| tr |
Türkisch |
| tt |
Tatarisch |
| uk |
Ukrainisch |
| ur |
Urdu |
| uz |
Usbekisch |
| vi |
Vietnamesisch |
Table MeasurementUnit_Enum
The measurement units, e.g. m
Table ParameterValue_Enum
Distinct values for a parameter of a method
Depending on:
Table PropertyType_Enum
The types of a Property, e.g. Chronostratigraphy
Dependent tables:
Table TaskDateType_Enum
The type of a task, e.g. freezing etc.
Dependent tables:
Table TaskModuleType_Enum
The type of a task, e.g. freezing etc.
Dependent tables:
Table TaskType_Enum
The type of a task, e.g. freezing etc.
Dependent tables:
Diversity Collection
VIEWS
The following objects are not included:
- System objects
- Objects marked as obsolete
- Previous versions of objects
View AnnotationEvent
Annotations linked to table CollectionEvent
| Column |
Data type |
Description |
Nullable |
| AnnotationID |
int |
ID of the annotation (primary key) |
NO |
| CollectionEventID |
int |
ID of the collection event. Refers to PK of table CollectionEvent |
NO |
| ReferencedAnnotationID |
int |
If an annotation refers to another annotation, the ID of the referred annotation |
YES |
| AnnotationType |
nvarchar (50) |
The type of the annotation as defined in AnnotationType_Enum, e.g. Reference |
NO |
| Title |
nvarchar (50) |
Title of the annotation |
YES |
| Annotation |
nvarchar (MAX) |
The annotation entered by the user |
NO |
| URI |
varchar (255) |
The complete URI address of a resource related to the annotation. May be link to a module, e.g. for the annotation type reference |
YES |
| ReferenceDisplayText |
nvarchar (500) |
The title of the reference. If the entry is linked to an external module like DiversityReferences, the cached display text of the referenced data set |
YES |
| ReferenceURI |
varchar (255) |
If the entry is linked to an external module like DiversityReferences, the link to the referenced data set |
YES |
| SourceDisplayText |
nvarchar (500) |
The name of the source. If the entry is linked to an external module like DiversityAgents, the cached display text of the referenced data set |
YES |
| SourceURI |
varchar (255) |
If the entry is linked to an external module like DiversityAgents, the link to the referenced data set |
YES |
| IsInternal |
bit |
If an annotation is restricted to authorized users of the database |
YES |
Depending on:
View AnnotationPart
Annotations linked to table CollectionSpecimenPart
| Column |
Data type |
Description |
Nullable |
| AnnotationID |
int |
ID of the annotation (primary key) |
NO |
| SpecimenPartID |
int |
ID of the collection specimen part. Refers to PK of table CollectionSpecimenPart |
NO |
| ReferencedAnnotationID |
int |
If an annotation refers to another annotation, the ID of the referred annotation |
YES |
| AnnotationType |
nvarchar (50) |
The type of the annotation as defined in AnnotationType_Enum, e.g. Reference |
NO |
| Title |
nvarchar (50) |
Title of the annotation |
YES |
| Annotation |
nvarchar (MAX) |
The annotation entered by the user |
NO |
| URI |
varchar (255) |
The complete URI address of a resource related to the annotation. May be link to a module, e.g. for the annotation type reference |
YES |
| ReferenceDisplayText |
nvarchar (500) |
The title of the reference. If the entry is linked to an external module like DiversityReferences, the cached display text of the referenced data set |
YES |
| ReferenceURI |
varchar (255) |
If the entry is linked to an external module like DiversityReferences, the link to the referenced data set |
YES |
| SourceDisplayText |
nvarchar (500) |
The name of the source. If the entry is linked to an external module like DiversityAgents, the cached display text of the referenced data set |
YES |
| SourceURI |
varchar (255) |
If the entry is linked to an external module like DiversityAgents, the link to the referenced data set |
YES |
| IsInternal |
bit |
If an annotation is restricted to authorized users of the database |
YES |
Depending on:
View AnnotationSpecimen
Annotations linked to table CollectionSpecimen
| Column |
Data type |
Description |
Nullable |
| AnnotationID |
int |
ID of the annotation (primary key) |
NO |
| CollectionSpecimenID |
int |
ID of the collection specimen. Refers to PK of table CollectionSpecimen |
NO |
| ReferencedAnnotationID |
int |
If an annotation refers to another annotation, the ID of the referred annotation |
YES |
| AnnotationType |
nvarchar (50) |
The type of the annotation as defined in AnnotationType_Enum, e.g. Reference |
NO |
| Title |
nvarchar (50) |
Title of the annotation |
YES |
| Annotation |
nvarchar (MAX) |
The annotation entered by the user |
NO |
| URI |
varchar (255) |
The complete URI address of a resource related to the annotation. May be link to a module, e.g. for the annotation type reference |
YES |
| ReferenceDisplayText |
nvarchar (500) |
The title of the reference. If the entry is linked to an external module like DiversityReferences, the cached display text of the referenced data set |
YES |
| ReferenceURI |
varchar (255) |
If the entry is linked to an external module like DiversityReferences, the link to the referenced data set |
YES |
| SourceDisplayText |
nvarchar (500) |
The name of the source. If the entry is linked to an external module like DiversityAgents, the cached display text of the referenced data set |
YES |
| SourceURI |
varchar (255) |
If the entry is linked to an external module like DiversityAgents, the link to the referenced data set |
YES |
| IsInternal |
bit |
If an annotation is restricted to authorized users of the database |
YES |
Depending on:
View AnnotationUnit
Annotations linked to table IdentificationUnit
| Column |
Data type |
Description |
Nullable |
| AnnotationID |
int |
ID of the annotation (primary key) |
NO |
| IdentificationUnitID |
int |
ID of the IdentificationUnit. Refers to PK of table IdentificationUnit |
NO |
| ReferencedAnnotationID |
int |
If an annotation refers to another annotation, the ID of the referred annotation |
YES |
| AnnotationType |
nvarchar (50) |
The type of the annotation as defined in AnnotationType_Enum, e.g. Reference |
NO |
| Title |
nvarchar (50) |
Title of the annotation |
YES |
| Annotation |
nvarchar (MAX) |
The annotation entered by the user |
NO |
| URI |
varchar (255) |
The complete URI address of a resource related to the annotation. May be link to a module, e.g. for the annotation type reference |
YES |
| ReferenceDisplayText |
nvarchar (500) |
The title of the reference. If the entry is linked to an external module like DiversityReferences, the cached display text of the referenced data set |
YES |
| ReferenceURI |
varchar (255) |
If the entry is linked to an external module like DiversityReferences, the link to the referenced data set |
YES |
| SourceDisplayText |
nvarchar (500) |
The name of the source. If the entry is linked to an external module like DiversityAgents, the cached display text of the referenced data set |
YES |
| SourceURI |
varchar (255) |
If the entry is linked to an external module like DiversityAgents, the link to the referenced data set |
YES |
| IsInternal |
bit |
If an annotation is restricted to authorized users of the database |
YES |
Depending on:
View CollectionAgent_Core
Table CollectionAgent restricted to available datasets
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Refers to ID of CollectionEvent (= foreign key and part of primary key) |
NO |
| CollectorsName |
nvarchar (255) |
Name of the Collector |
NO |
| CollectorsAgentURI |
varchar (255) |
The URI of the Agent, e.g. as stored within the module DiversityAgents |
YES |
| CollectorsSequence |
datetime2 |
The order of collectors in a team. Automatically set by the database system |
YES |
| CollectorsNumber |
nvarchar (50) |
Number assigned to a specimen or a batch of specimens by the collector during the collection event (= ‘field number’) |
YES |
| Notes |
nvarchar (MAX) |
Notes on the collector, e.g. if the name is uncertain |
YES |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
Depending on:
- CollectionAgent
- CollectionSpecimenID_Available
View CollectionEvent_Core2
List for accessible content of table CollectionEvent with preformated date column
| Column |
Data type |
Description |
Nullable |
| CollectionEventID |
int |
Unique ID for the table CollectionEvent (= primary key) |
NO |
| Version |
int |
The version of the data set. Automatically set by the system. |
NO |
| SeriesID |
int |
The ID of the related expedition. Relates to the PK of the table CollectionExpedition (foreign key). |
YES |
| CollectorsEventNumber |
nvarchar (50) |
Number assigned to a collection event by the collector (= ‘field number’) |
YES |
| CollectionDate |
datetime |
The cached date of the collection event calulated from the entries in CollectionDay, -Month and -Year. |
YES |
| CollectionDay |
tinyint |
The day of the date of the event or when the collection event started |
YES |
| CollectionMonth |
tinyint |
The month of the date of the event or when the collection event started |
YES |
| CollectionYear |
smallint |
The year of the date of the event or when the collection event started |
YES |
| CollectionEndDay |
tinyint |
The day of the date of the event or when the collection event ended |
YES |
| CollectionEndMonth |
tinyint |
The month of the date of the event or when the collection event ended |
YES |
| CollectionEndYear |
smallint |
The year of the date of the event or when the collection event ended |
YES |
| CollectionDateSupplement |
nvarchar (100) |
Verbal or additional collection date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. The end date, if the collection event comprises a period. The time of the event, if necessary. |
YES |
| CollectionDateCategory |
nvarchar (50) |
Category of the date of the identification e.g. “system”, “estimated” (= foreign key, see in table CollEventDateCategory_Enum) |
YES |
| CollectionTime |
varchar (50) |
The time of the event or when the collection event started |
YES |
| CollectionTimeSpan |
varchar (50) |
The time span e.g. in seconds of the collection event |
YES |
| LocalityDescription |
nvarchar (MAX) |
Locality description of the locality exactly as written on the original label (i.e. without corrections during data entry) |
YES |
| LocalityVerbatim |
nvarchar (MAX) |
Locality as given in historical context, documents and labels |
YES |
| HabitatDescription |
nvarchar (MAX) |
Geo-ecological description of the locality exactly as written on the original label (i.e. without corrections during data entry) |
YES |
| ReferenceTitle |
nvarchar (255) |
The title of the publication where the collection event was published. Note that this is only a cached value where ReferenceURI is present |
YES |
| ReferenceDetails |
nvarchar (50) |
The exact location within the reference, e.g. pages, plates |
YES |
| ReferenceURI |
varchar (255) |
URI (e.g. LSID) of the source publication where the collection event is published, may e.g. refer to the module DiversityReferences |
YES |
| CollectingMethod |
nvarchar (MAX) |
Description of the method used for collecting the samples, e.g. traps, moist chambers, drag net |
YES |
| Notes |
nvarchar (MAX) |
Notes on the collection event |
YES |
| CountryCache |
nvarchar (50) |
The country where the collection event took place. Cached value derived from an entry in CollectionEventLocalisation |
YES |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
| DataWithholdingReasonDate |
nvarchar (50) |
The reason for withholding the collection date |
YES |
| CollectionDate_YMD |
varchar (94) |
- |
YES |
Depending on:
- CollectionEvent
- CollectionEventID_AvailableNotReadOnly
View CollectionEventID_CanEdit
List for CollectionEventID (PK of table CollectionEvent) a user can edit
| Column |
Data type |
Description |
Nullable |
| CollectionEventID |
int |
Refers to the ID of table CollectionEvent (= foreign key and part of primary key) |
YES |
Depending on:
- CollectionProject
- CollectionSpecimen
- ProjectUser
View CollectionEventID_UserAvailable
List for CollectionEventID (PK of table CollectionEvent) a user has access to
| Column |
Data type |
Description |
Nullable |
| CollectionEventID |
int |
Refers to the ID of table CollectionEvent (= foreign key and part of primary key) |
YES |
Depending on:
- CollectionProject
- CollectionSpecimen
- ProjectUser
View CollectionEventLocalisation_Core
Content of table CollectionEventLocalisation excluding log columns and geography
| Column |
Data type |
Description |
Nullable |
| CollectionEventID |
int |
Refers to the ID of CollectionEvent (= foreign key and part of primary key) |
NO |
| LocalisationSystemID |
int |
Refers to the ID of LocalisationSystem (= foreign key and part of primary key) |
NO |
| Location1 |
nvarchar (255) |
Either a named location selected from a thesaurus (e.g. ‘Germany, Bavaria, Kleindingharting’) or altitude range or other values (e. g. 100-200 m) |
YES |
| Location2 |
nvarchar (255) |
Corresponding value to Location1 e.g. ID or URI of gazetteer or thesaurus |
YES |
| LocationAccuracy |
nvarchar (50) |
The accuracy of the determination of this locality |
YES |
| LocationNotes |
nvarchar (MAX) |
Notes on the location |
YES |
| DeterminationDate |
smalldatetime |
Date of the determination of the geographical localisation |
YES |
| DistanceToLocation |
varchar (50) |
Distance from the specified place to the real location of the collection site (m) |
YES |
| DirectionToLocation |
varchar (50) |
Direction from the specified place to the real location of the collection site (Degrees rel. to north) |
YES |
| ResponsibleName |
nvarchar (255) |
The name of the agent (person or organization) responsible for this entry. |
YES |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or organisation responsible for the data (see e.g. module DiversityAgents) |
YES |
| AverageAltitudeCache |
float |
Calculated altitude as parsed from the location fields |
YES |
| AverageLatitudeCache |
float |
Calculated latitude as parsed from the location fields |
YES |
| AverageLongitudeCache |
float |
Calculated longitude as parsed from the location fields |
YES |
| RecordingMethod |
nvarchar (500) |
The method or device used for the recording of the localisation |
YES |
| Geography |
nvarchar (MAX) |
The geography of the localisation |
YES |
Depending on:
- CollectionEventLocalisation
View CollectionSpecimen_Core2
List of accessible dataset of table CollectionSpecimen containing additional data from table CollectionEvent
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Unique ID for the table CollectionSpecimen (primary key) |
NO |
| Version |
int |
The version of the data set. Automatically set by the system. |
NO |
| CollectionEventID |
int |
Unique ID for the table CollectionEvent (= primary key) |
YES |
| AccessionNumber |
nvarchar (50) |
Accession number of the specimen within the collection, e.g. “M-29834752” |
YES |
| AccessionDate |
datetime |
The date of the accession calculated from the entries in AccessionDay, -Month and -Year |
YES |
| AccessionDay |
tinyint |
The day of the date when the specimen was acquired in the collection |
YES |
| AccessionMonth |
tinyint |
The month of the date when the specimen was acquired in the collection |
YES |
| AccessionYear |
smallint |
The year of the date when the specimen was acquired in the collection |
YES |
| AccessionDateSupplement |
nvarchar (255) |
Verbal or additional accession date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
YES |
| AccessionDateCategory |
nvarchar (50) |
Category of the date of the accession e.g. “system”, “estimated” (= foreign key, see in table CollDateCategory_Enum) |
YES |
| DepositorsName |
nvarchar (255) |
The name of the depositor(s) (person or organization responsible for deposition). Where entire collections are deposited, this should also contain the collection name (e.g. ‘Herbarium P. Döbbler’) |
YES |
| DepositorsAgentURI |
varchar (255) |
The URI of the depositor(s) (person or organization responsible for deposition) |
YES |
| DepositorsAccessionNumber |
nvarchar (50) |
Accession number of the specimen within the previous or original collection, e.g. ‘D-23948’ |
YES |
| LabelTitle |
nvarchar (MAX) |
The title of the label e.g. for printing labels. |
YES |
| LabelType |
nvarchar (50) |
Printed, typewritten, typewritten with handwriting added, entirely in handwriting, etc. |
YES |
| LabelTranscriptionState |
nvarchar (50) |
The state of the transcription of a label into the database: ‘Not started’, ‘incomplete’, ‘complete’ |
YES |
| LabelTranscriptionNotes |
nvarchar (255) |
User defined notes on the transcription of the label into the database |
YES |
| ExsiccataURI |
varchar (255) |
If specimen is an exsiccata: The URI of the exsiccata series, e.g. as stored within the DiversityExsiccata module |
YES |
| ExsiccataAbbreviation |
nvarchar (255) |
If specimen is an exsiccata: Standard abbreviation of the exsiccata (not necessarily a unique identifier; editors or publication places may change over time) |
YES |
| OriginalNotes |
nvarchar (MAX) |
Notes found on the label of the specimen by the original collector or from a later revision |
YES |
| AdditionalNotes |
nvarchar (MAX) |
Additional notes made by the editor of the specimen record, e.g. ‘doubtful identification/locality’ |
YES |
| ReferenceTitle |
nvarchar (255) |
The title of the publication where the collection event was published. Note that this is only a cached value where ReferenceURI is present |
YES |
| ReferenceURI |
varchar (255) |
URI (e.g. LSID) of the source publication where the collection event is published, may e.g. refer to the module DiversityReferences |
YES |
| Problems |
nvarchar (255) |
Description of a problem which occurred during data editing. Typically these entries should be deleted after help has been obtained. Do not enter scientific problems here; use AdditionalNotes for such permanent problems! |
YES |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
| CollectionDate |
varchar (94) |
The cached date of the collection event calulated from the entries in CollectionDay, -Month and -Year. |
YES |
| Locality |
nvarchar (MAX) |
LocalityDescription from table CollectionEvent |
YES |
| Habitat |
nvarchar (MAX) |
HabitatDescription from table CollectionEvent |
YES |
| InternalNotes |
nvarchar (MAX) |
Internal notes which should not be published e.g. on websites |
YES |
| ExternalDatasourceID |
int |
An ID to identify an external data collection of the collected specimen (primary key, the ID has no meaning outside of the DiversityWorkbench system) |
YES |
| ExternalIdentifier |
nvarchar (100) |
The identifier of the external specimen as defined in the external data source |
YES |
| LogCreatedWhen |
datetime |
Point in time when this data set was created |
YES |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set |
YES |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last |
YES |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last |
YES |
Depending on:
- CollectionEvent
- CollectionSpecimen
- CollectionSpecimenID_Available
- UserProxy
View CollectionSpecimenID_Available
CollectionSpecimenIDs a user has access to including CollectionSpecimenIDs not linked to a project
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Depending on:
- CollectionProject
- CollectionSpecimen
- ProjectUser
View CollectionSpecimenID_AvailableReadOnly
CollectionSpecimenIDs a user has read only access to including CollectionSpecimenIDs from locked projects
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Depending on:
- CollectionProject
- CollectionSpecimenID_Locked
- ProjectUser
View CollectionSpecimenID_CanEdit
CollectionSpecimenIDs a user has access to excluding those with read only access
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Depending on:
- CollectionProject
- ProjectUser
View CollectionSpecimenID_Locked
CollectionSpecimenIDs of locked projects
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Depending on:
- CollectionProject
- ProjectProxy
View CollectionSpecimenID_ReadOnly
CollectionSpecimenIDs a user has read only access to including CollectionSpecimenIDs from locked projects
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Depending on:
- CollectionProject
- CollectionSpecimenID_Locked
- ProjectUser
View CollectionSpecimenID_UserAvailable
CollectionSpecimenIDs a user has access to including CollectionSpecimenIDs not linked to a project
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Depending on:
- CollectionProject
- CollectionSpecimen
- ProjectUser
View CollectionSpecimenPart_Core2
Content of table CollectionSpecimenPart restricted to those avaliable for a user including the collection hierarchy
| Column |
Data type |
Description |
Nullable |
| Collection |
nvarchar (1158) |
Hierarchy of the collection separated by " |
" followed by StorageLocation resp. AccessionNumber and Sublabel |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen (= part of primary key). |
NO |
| DerivedFromSpecimenPartID |
int |
SpecimenPartID of the specimen from which the current specimen is derived from |
YES |
| PreparationMethod |
nvarchar (MAX) |
The method used for the preparation of the part of the specimen, e.g. the inoculation method for cultures |
YES |
| PreparationDate |
datetime |
Point in time when the part was preparated e.g when it was separated from the source object |
YES |
| AccessionNumber |
nvarchar (50) |
Accession number of the part of the specimen within the collection, if it is different from the accession number of the specimen as stored in the table CollectionSpecimen, e.g. “M-29834752” |
YES |
| PartSublabel |
nvarchar (50) |
The label for a part of a specimen, e.g. “cone”, or a number attached to a duplicate of a specimen |
YES |
| CollectionID |
int |
ID of the collection as stored in table Collection (= foreign key, see table Collection) |
NO |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. (= foreign key, see table CollMaterialCategory_Enum) |
NO |
| StorageLocation |
nvarchar (255) |
A code identifying the place where the specimen is stored within the collection. Frequently the accepted scientific name is used as storage location code. |
YES |
| StorageContainer |
nvarchar (500) |
The container in which the part is stored |
YES |
| Stock |
float |
Number of stock units, if the specimen is stored in separated units e.g. several boxes or vessels (max. 255) |
YES |
| StockUnit |
nvarchar (50) |
If empty, the stock is given as a count, else it contains the unit in which stock is expressed, e.g. µl, ml, kg etc. |
YES |
| Notes |
nvarchar (MAX) |
Notes on the storage of the sample |
YES |
| PartAccessionNumber |
nvarchar (50) |
- |
YES |
Depending on:
- CollectionHierarchyAll
- CollectionSpecimenID_UserAvailable
- CollectionSpecimenPart
View CollectionSpecimenRelationInternal
List of specimen with a relation to a specimen within the same database or where another specimen within the database has defined a relation to
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Unique ID for the table CollectionSpecimen (primary key) |
YES |
| RelatedSpecimen_CollectionSpecimenID |
int |
CollectionSpecimenID of the related specimen |
YES |
| RelatedSpecimenDisplayText |
nvarchar (255) |
The name of a related specimen as shown e.g. in a user interface |
YES |
| RelationType |
nvarchar (50) |
Type of the relation between the specimen (= foreign key, see table CollRelationType_Enum) |
YES |
| RelatedSpecimenCollectionID |
int |
ID of the Collection as stored in table Collection (= foreign key, see table Collection) |
YES |
| RelatedSpecimenDescription |
nvarchar (MAX) |
Description of the related specimen |
YES |
| Notes |
nvarchar (MAX) |
Notes on the relation to the specimen |
YES |
Depending on:
- baseURL
- CollectionSpecimen
- CollectionSpecimenRelation
View CollectionSpecimenRelationInvers
Content of function CollectionSpecimenRelationInversList: List of specimen where another specimen within the database has defined a relation to
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Unique ID for the table CollectionSpecimen (primary key) |
NO |
| RelatedSpecimenCollectionSpecimenID |
int |
The CollectionSpecimenID of the dataset that refers to the current dataset |
NO |
| RelatedSpecimenAccessionNumber |
nvarchar (50) |
The AccessionNumber of the dataset that refers to the current dataset |
YES |
| RelationType |
nvarchar (50) |
Type of the relation between the specimen (= foreign key, see table CollRelationType_Enum) |
YES |
| RelatedSpecimenCollectionID |
int |
ID of the Collection as stored in table Collection (= foreign key, see table Collection) |
YES |
| Notes |
nvarchar (MAX) |
Notes on the relation to the specimen |
YES |
Depending on:
- CollectionSpecimenRelationInversList
View CollectionSpecimenTransactionRequest
PK and supplementary columns of Table CollectionSpecimenTransaction with TransactionType loan or request the user has access to as a CollectionManager or the user has created and access to as a CollectionRequester
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Refers to ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
| TransactionID |
int |
Unique ID for the table Transaction (= foreign key and part of primary key) |
NO |
| SpecimenPartID |
int |
Optional: If the data set is related to a part of a specimen, the ID of a related part (= foreign key, see table CollectionSpecimenPart) |
NO |
| IsOnLoan |
bit |
True, if a specimen is on loan |
YES |
| LogInsertedBy |
nvarchar (50) |
Name of user to first enter (typ or import) the data. |
YES |
| LogInsertedWhen |
smalldatetime |
Point in time when the data was first entered (typed or imported) into this database. |
YES |
Depending on:
- CollectionSpecimenTransaction
- TransactionRequest
View FirstLinesIdentification
Content of table Identification restricted to last identification of a IdentificationUnit
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
| IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
NO |
| IdentificationSequence |
smallint |
The sequence of the identifications. The last identification (having the highest sequence) is regarded as valid |
NO |
| IdentificationDate |
datetime |
The date of the identification calculated from the entries in IdentificationDay, -Month and -Year |
YES |
| IdentificationDay |
tinyint |
The day of the identification |
YES |
| IdentificationMonth |
tinyint |
The month of the identification |
YES |
| IdentificationYear |
smallint |
The year of the identification. The year may be empty if only the day or month are known. |
YES |
| IdentificationDateSupplement |
nvarchar (255) |
Verbal or additional identification date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
YES |
| IdentificationDateCategory |
nvarchar (50) |
Category of the date of the identification e.g. “system”, “estimated” (= foreign key, see in table CollDateCategory_Enum) |
YES |
| VernacularTerm |
nvarchar (255) |
Name or term other than a taxonomic (= scientific) name, e.g. ‘pine’, ’limestone’, ‘conifer’, ‘hardwood’ |
YES |
| TaxonomicName |
nvarchar (255) |
Valid name of the species (including the taxonomic author where available). Example: ‘Rosa canina L.’ |
YES |
| NameURI |
varchar (255) |
The URI of the taxonomic name, e.g. as provided by the module DiversityTaxonNames. |
YES |
| IdentificationCategory |
nvarchar (50) |
Category of the identification e.g. ‘determination’, ‘confirmation’, ‘absence’ (= foreign key, see table CollIdentificationCategory_Enum) |
YES |
| IdentificationQualifier |
nvarchar (50) |
Qualification of the identification e.g. “cf.”," aff.", “sp. nov.” (= foreign key, see table CollIdentificationQualifier_Enum) |
YES |
| TypeStatus |
nvarchar (50) |
If identification unit is type of a taxonomic name: holotype, syntype, etc. (= foreign key, see table CollTypeStatus_Enum) |
YES |
| TypeNotes |
nvarchar (MAX) |
Notes on the typification of this specimen |
YES |
| Notes |
nvarchar (MAX) |
User defined notes, e.g. the reason for a re-determination / change of the name, etc. |
YES |
| ResponsibleName |
nvarchar (255) |
Name of the person or institution responsible for the determination |
YES |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
YES |
| LogCreatedWhen |
datetime |
Point in time when this data set was created |
YES |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set |
YES |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last |
YES |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last |
YES |
Depending on:
- FirstLinesIdentificationUnit
- Identification
View Identification_Core2
Content of table Identification restricted to those available for a user
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
| IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
NO |
| IdentificationSequence |
smallint |
The sequence of the identifications. The last identification (having the highest sequence) is regarded as valid |
NO |
| IdentificationDate |
datetime |
The date of the identification calculated from the entries in IdentificationDay, -Month and -Year |
YES |
| IdentificationDay |
tinyint |
The day of the identification |
YES |
| IdentificationMonth |
tinyint |
The month of the identification |
YES |
| IdentificationYear |
smallint |
The year of the identification. The year may be empty if only the day or month are known. |
YES |
| IdentificationDateSupplement |
nvarchar (255) |
Verbal or additional identification date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
YES |
| IdentificationDateCategory |
nvarchar (50) |
Category of the date of the identification e.g. “system”, “estimated” (= foreign key, see in table CollDateCategory_Enum) |
YES |
| VernacularTerm |
nvarchar (255) |
Name or term other than a taxonomic (= scientific) name, e.g. ‘pine’, ’limestone’, ‘conifer’, ‘hardwood’ |
YES |
| TaxonomicName |
nvarchar (255) |
Valid name of the species (including the taxonomic author where available). Example: ‘Rosa canina L.’ |
YES |
| NameURI |
varchar (255) |
The URI of the taxonomic name, e.g. as provided by the module DiversityTaxonNames. |
YES |
| IdentificationCategory |
nvarchar (50) |
Category of the identification e.g. ‘determination’, ‘confirmation’, ‘absence’ (= foreign key, see table CollIdentificationCategory_Enum) |
YES |
| IdentificationQualifier |
nvarchar (50) |
Qualification of the identification e.g. “cf.”," aff.", “sp. nov.” (= foreign key, see table CollIdentificationQualifier_Enum) |
YES |
| TypeStatus |
nvarchar (50) |
If identification unit is type of a taxonomic name: holotype, syntype, etc. (= foreign key, see table CollTypeStatus_Enum) |
YES |
| TypeNotes |
nvarchar (MAX) |
Notes on the typification of this specimen |
YES |
| Notes |
nvarchar (MAX) |
User defined notes, e.g. the reason for a re-determination / change of the name, etc. |
YES |
| ResponsibleName |
nvarchar (255) |
Name of the person or institution responsible for the determination |
YES |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
YES |
Depending on:
- CollectionSpecimenID_Available
- Identification
View IdentificationUnit_Core2
Content of table IdentificationUnit restricted to those available for a user
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
| IdentificationUnitID |
int |
ID of the IdentificationUnit (= part of primary key). Usually one of possibly several organisms present on the collected specimen. Example: parasite with hyperparasite on plant leaf = 3 units, |
NO |
| LastIdentificationCache |
nvarchar (255) |
The last identification as entered in table Identification |
NO |
| FamilyCache |
nvarchar (255) |
A cached value of the family of the taxon of the last identification. Can be set by the editor, if NameURI in table Identification is NULL, otherwise set by the system. |
YES |
| OrderCache |
nvarchar (255) |
A cached value of the order of the taxon of the last identification. Can be set by the editor, if NameURI in table Identification is NULL, otherwise set by the system. |
YES |
| TaxonomicGroup |
nvarchar (50) |
Taxonomic group the organism, identified by this unit, belongs to. Groups listed in table CollTaxonomicGroup_Enum (= foreign key) |
NO |
| OnlyObserved |
bit |
True, if the organism was only observed rather than collected. It is therefore not present on the preserved specimen. Example: Tree under which the collected mycorrhizal fungus grew. |
YES |
| RelatedUnitID |
int |
The IdentificationUnitID of the organism or substrate on which this organism is growing (= foreign key) |
YES |
| RelationType |
nvarchar (50) |
The relation of a unit to its substrate, e.g. parasitism, symbiosis etc. as stored in CollRelationType_Enum (= foreign key) |
YES |
| ColonisedSubstratePart |
nvarchar (255) |
If a substrate association exists: part of the substrate which is affected in the interaction (e.g. ’leaves’, if a fungus is growing on the leaves of an infected plant) |
YES |
| LifeStage |
nvarchar (255) |
Examples: ‘II, III’ for spore generations of rusts or ‘seed’, ‘seedling’ etc. for higher plants |
YES |
| Gender |
nvarchar (50) |
The sex of the organism, e.g. ‘female’ |
YES |
| NumberOfUnits |
smallint |
The number of units of this organism, e.g. 400 beetles in a bottle |
YES |
| ExsiccataNumber |
nvarchar (50) |
If specimen is an exsiccata: Number of current specimen within the exsiccata series |
YES |
| ExsiccataIdentification |
smallint |
Refers to the IdentificationSequence in Identification (= foreign key). The name under which the collectied specimen or this organism is published within an exsiccata. |
YES |
| UnitIdentifier |
nvarchar (50) |
An identifier for the identification of the unit, e.g. a number painted on a tree within an experimental plot |
YES |
| UnitDescription |
nvarchar (50) |
Description of the unit, especially if not an organism but parts or remnants of it were present or observed, e.g. a nest of an insect or a song of a bird |
YES |
| Circumstances |
nvarchar (50) |
Circumstances of the occurence of the organism |
YES |
| DisplayOrder |
smallint |
The sequence in which the units within this specimen will appear on e.g. a label where the first unit may be printed in the header and others in the text below. 0 means the unit should not appear on a label. |
NO |
| Notes |
nvarchar (MAX) |
Further information on the organism or interaction, e.g. infection symptoms like ‘producing galls’ |
YES |
| HierarchyCache |
nvarchar (500) |
A cached value fo the superior taxonomy of the last identification as derived from a taxonomic data provider |
YES |
| RetrievalType |
nvarchar (50) |
The way the data about the unit were retrieved, e.g. observation, literature |
YES |
| ParentUnitID |
int |
The IdentificationUnitID of a parent organism of which this organism is a child of (= foreign key). |
YES |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
| NumberOfUnitsModifier |
nvarchar (50) |
A modifier for the number of units of this organism, e.g. ca. 400 beetles in a bottle |
YES |
Depending on:
- CollectionSpecimenID_Available
- IdentificationUnit
View IdentificationUnitDisplayOrder1
Content of table IdentificationUnit restricted to those available for a user and DisplayOrder = 1
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
| IdentificationUnitID |
int |
ID of the IdentificationUnit (= part of primary key). Usually one of possibly several organisms present on the collected specimen. Example: parasite with hyperparasite on plant leaf = 3 units, |
NO |
| LastIdentificationCache |
nvarchar (255) |
The last identification as entered in table Identification |
NO |
| FamilyCache |
nvarchar (255) |
A cached value of the family of the taxon of the last identification. Can be set by the editor, if NameURI in table Identification is NULL, otherwise set by the system. |
YES |
| OrderCache |
nvarchar (255) |
A cached value of the order of the taxon of the last identification. Can be set by the editor, if NameURI in table Identification is NULL, otherwise set by the system. |
YES |
| TaxonomicGroup |
nvarchar (50) |
Taxonomic group the organism, identified by this unit, belongs to. Groups listed in table CollTaxonomicGroup_Enum (= foreign key) |
NO |
| OnlyObserved |
bit |
True, if the organism was only observed rather than collected. It is therefore not present on the preserved specimen. Example: Tree under which the collected mycorrhizal fungus grew. |
YES |
| RelatedUnitID |
int |
The IdentificationUnitID of the organism or substrate on which this organism is growing (= foreign key) |
YES |
| RelationType |
nvarchar (50) |
The relation of a unit to its substrate, e.g. parasitism, symbiosis etc. as stored in CollRelationType_Enum (= foreign key) |
YES |
| ColonisedSubstratePart |
nvarchar (255) |
If a substrate association exists: part of the substrate which is affected in the interaction (e.g. ’leaves’, if a fungus is growing on the leaves of an infected plant) |
YES |
| LifeStage |
nvarchar (255) |
Examples: ‘II, III’ for spore generations of rusts or ‘seed’, ‘seedling’ etc. for higher plants |
YES |
| Gender |
nvarchar (50) |
The sex of the organism, e.g. ‘female’ |
YES |
| NumberOfUnits |
smallint |
The number of units of this organism, e.g. 400 beetles in a bottle |
YES |
| ExsiccataNumber |
nvarchar (50) |
If specimen is an exsiccata: Number of current specimen within the exsiccata series |
YES |
| ExsiccataIdentification |
smallint |
Refers to the IdentificationSequence in Identification (= foreign key). The name under which the collectied specimen or this organism is published within an exsiccata. |
YES |
| UnitIdentifier |
nvarchar (50) |
An identifier for the identification of the unit, e.g. a number painted on a tree within an experimental plot |
YES |
| Circumstances |
nvarchar (50) |
Circumstances of the occurence of the organism |
YES |
| DisplayOrder |
smallint |
The sequence in which the units within this specimen will appear on e.g. a label where the first unit may be printed in the header and others in the text below. 0 means the unit should not appear on a label. |
NO |
| Notes |
nvarchar (MAX) |
Further information on the organism or interaction, e.g. infection symptoms like ‘producing galls’ |
YES |
Depending on:
- CollectionSpecimenID_UserAvailable
- IdentificationUnit
View ManagerSpecimenPartList
List based on content of table CollectionSpecimenPart restricted to those with an AccessionNumber and available for a CollectionManager
| Column |
Data type |
Description |
Nullable |
| AccessionNumber |
nvarchar (50) |
Accession number of the specimen within the collection, e.g. “M-29834752” |
YES |
| CollectionSpecimenID |
int |
Unique ID for the table CollectionSpecimen (primary key) |
NO |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen (= part of primary key). |
NO |
| AccessionNumberSpecimen |
nvarchar (50) |
- |
YES |
| AccessionNumberPart |
nvarchar (50) |
- |
YES |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. (= foreign key, see table CollMaterialCategory_Enum) |
NO |
| CollectionName |
nvarchar (50) |
- |
YES |
Depending on:
- CollectionSpecimen
- CollectionSpecimenPart
- ManagerCollectionList
View ProjectList
List of projects available for a user
| Column |
Data type |
Description |
Nullable |
| Project |
nvarchar (50) |
The name or title of the project as shown in a user interface (Projects are defined in DiversityProjects) |
YES |
| ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
NO |
| ReadOnly |
int |
If the user has only read access to data of this project |
YES |
| ProjectURI |
varchar (255) |
The URI of the project, e.g. as provided by the module DiversityProjects. |
YES |
Depending on:
View ProjectListNotReadOnly
List of projects a user can edit
| Column |
Data type |
Description |
Nullable |
| Project |
nvarchar (50) |
The name or title of the project as shown in a user interface (Projects are defined in DiversityProjects) |
YES |
| ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
NO |
Depending on:
View RequesterSpecimenPartList
List of possible request for a user based on table CollectionSpecimenPart
| Column |
Data type |
Description |
Nullable |
| AccessionNumber |
nvarchar (50) |
Accession number of the specimen within the collection, e.g. “M-29834752” |
YES |
| CollectionSpecimenID |
int |
Unique ID for the table CollectionSpecimen (primary key) |
NO |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen (= part of primary key). |
NO |
| AccessionNumberSpecimen |
nvarchar (50) |
- |
YES |
| AccessionNumberPart |
nvarchar (50) |
- |
YES |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. (= foreign key, see table CollMaterialCategory_Enum) |
NO |
| CollectionName |
nvarchar (50) |
- |
YES |
Depending on:
- CollectionSpecimen
- CollectionSpecimenPart
- RequesterCollectionList
View TransactionForeignRequest
Content of table Transaction with TransactionType = request a CollectionManager has access to
| Column |
Data type |
Description |
Nullable |
| TransactionID |
int |
Unique ID for the transaction (= primary key) |
NO |
| ParentTransactionID |
int |
The ID of a preceeding transaction of a superior transaction, if transactions are organized in a hierarchy |
YES |
| TransactionType |
nvarchar (50) |
Type of the transaction, e.g. gift in or out, exchange in or out, purchase in or out |
NO |
| TransactionTitle |
nvarchar (200) |
The title of the transaction as e.g. shown in an user interface |
NO |
| ReportingCategory |
nvarchar (50) |
A group defined for the transaction, e.g. a taxonomic group as used for exchange balancing |
YES |
| AdministratingCollectionID |
int |
ID for the collection for which the Manager has the right to administrate the transaction. Corresponds to AdministratingCollectionID in table Transaction. |
NO |
| MaterialDescription |
nvarchar (MAX) |
Description of the material of this transaction |
YES |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. |
YES |
| MaterialCollectors |
nvarchar (MAX) |
The collectors of the material |
YES |
| FromCollectionID |
int |
The ID of the collection from which the specimen were transfered, e.g. the donating collection of a gift |
YES |
| FromTransactionPartnerName |
nvarchar (255) |
Name of the person or institution from which the specimen were transfered, e.g. the donator of a gift |
YES |
| FromTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
YES |
| FromTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the source of the specimen, e.g. the donating collection of a gift |
YES |
| ToCollectionID |
int |
The ID of the collection to which the specimen were transfered, e.g. the receiver of a gift |
YES |
| ToTransactionPartnerName |
nvarchar (255) |
Name of the person or institution to which the specimen were transfered, e.g. the receiver of a gift |
YES |
| ToTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
YES |
| ToTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the destination of the specimen, e.g. the receiving collection of a gift |
YES |
| NumberOfUnits |
smallint |
The number of units which were (initially) included in the transaction |
YES |
| Investigator |
nvarchar (50) |
The investigator for whose study a transacted material was sent |
YES |
| TransactionComment |
nvarchar (MAX) |
Comments on the exchanged material addressed to the transaction partner |
YES |
| BeginDate |
datetime |
Date when the transaction started |
YES |
| AgreedEndDate |
datetime |
End of the transaction period, e.g. if the time for borrowing the specimen is restricted |
YES |
| ActualEndDate |
datetime |
Actual end of the transaction after a prolonation when e.g. the date of return for a loan was prolonged by the owner |
YES |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction not to be published e.g. on a web page |
YES |
| ResponsibleName |
nvarchar (255) |
The person responsible for this transaction |
YES |
| ResponsibleAgentURI |
varchar (255) |
The URI of the person, team or organisation responsible for the data (see e.g. module DiversityAgents) |
YES |
Depending on:
- CollectionManager
- Transaction
View TransactionList
Content of table Transaction missing a AdministratingCollectionID resp. those available for a CollectionManager
| Column |
Data type |
Description |
Nullable |
| TransactionID |
int |
Unique ID for the transaction (= primary key) |
NO |
| ParentTransactionID |
int |
The ID of a preceeding transaction of a superior transaction, if transactions are organized in a hierarchy |
YES |
| TransactionType |
nvarchar (50) |
Type of the transaction, e.g. gift in or out, exchange in or out, purchase in or out |
NO |
| TransactionTitle |
nvarchar (200) |
The title of the transaction as e.g. shown in an user interface |
NO |
| ReportingCategory |
nvarchar (50) |
A group defined for the transaction, e.g. a taxonomic group as used for exchange balancing |
YES |
| AdministratingCollectionID |
int |
ID for the collection for which the Manager has the right to administrate the transaction. Corresponds to AdministratingCollectionID in table Transaction. |
NO |
| MaterialDescription |
nvarchar (MAX) |
Description of the material of this transaction |
YES |
| MaterialSource |
nvarchar (500) |
The source of the material within a transaction, e.g. a excavation |
YES |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. |
YES |
| MaterialCollectors |
nvarchar (MAX) |
The collectors of the material |
YES |
| FromCollectionID |
int |
The ID of the collection from which the specimen were transfered, e.g. the donating collection of a gift |
YES |
| FromTransactionPartnerName |
nvarchar (255) |
Name of the person or institution from which the specimen were transfered, e.g. the donator of a gift |
YES |
| FromTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
YES |
| FromTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the source of the specimen, e.g. the donating collection of a gift |
YES |
| ToCollectionID |
int |
The ID of the collection to which the specimen were transfered, e.g. the receiver of a gift |
YES |
| ToTransactionPartnerName |
nvarchar (255) |
Name of the person or institution to which the specimen were transfered, e.g. the receiver of a gift |
YES |
| ToTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
YES |
| ToTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the destination of the specimen, e.g. the receiving collection of a gift |
YES |
| ToRecipient |
nvarchar (255) |
The recipient receiving the transaction e.g. if not derived from the link to DiversityAgents |
YES |
| NumberOfUnits |
smallint |
The number of units which were (initially) included in the transaction |
YES |
| Investigator |
nvarchar (200) |
The investigator for whose study a transacted material was sent |
YES |
| TransactionComment |
nvarchar (MAX) |
Comments on the exchanged material addressed to the transaction partner |
YES |
| BeginDate |
datetime |
Date when the transaction started |
YES |
| AgreedEndDate |
datetime |
End of the transaction period, e.g. if the time for borrowing the specimen is restricted |
YES |
| ActualEndDate |
datetime |
Actual end of the transaction after a prolonation when e.g. the date of return for a loan was prolonged by the owner |
YES |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction not to be published e.g. on a web page |
YES |
| ResponsibleName |
nvarchar (255) |
The person responsible for this transaction |
YES |
| ResponsibleAgentURI |
varchar (255) |
The URI of the person, team or organisation responsible for the data (see e.g. module DiversityAgents) |
YES |
| DateSupplement |
nvarchar (100) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. |
YES |
Depending on:
- CollectionManager
- Transaction
View TransactionList_H7
Content of table Transaction missing a AdministratingCollectionID resp. those available for a CollectionManager including a hierarchy with up to 7 levels
| Column |
Data type |
Description |
Nullable |
| TransactionID |
int |
Unique ID for the transaction (= primary key) |
NO |
| ParentTransactionID |
int |
The ID of a preceeding transaction of a superior transaction, if transactions are organized in a hierarchy |
YES |
| TransactionType |
nvarchar (50) |
Type of the transaction, e.g. gift in or out, exchange in or out, purchase in or out |
NO |
| TransactionTitle |
nvarchar (200) |
The title of the transaction as e.g. shown in an user interface |
NO |
| HierarchyDisplayText |
nvarchar (1621) |
Hierarchy of the transaction represented by the TransactionTitle separated by " |
" for up to 7 levels |
| ReportingCategory |
nvarchar (50) |
A group defined for the transaction, e.g. a taxonomic group as used for exchange balancing |
YES |
| AdministratingCollectionID |
int |
ID for the collection for which the Manager has the right to administrate the transaction. Corresponds to AdministratingCollectionID in table Transaction. |
NO |
| MaterialDescription |
nvarchar (MAX) |
Description of the material of this transaction |
YES |
| MaterialSource |
nvarchar (500) |
The source of the material within a transaction, e.g. a excavation |
YES |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. |
YES |
| MaterialCollectors |
nvarchar (MAX) |
The collectors of the material |
YES |
| FromCollectionID |
int |
The ID of the collection from which the specimen were transfered, e.g. the donating collection of a gift |
YES |
| FromTransactionPartnerName |
nvarchar (255) |
Name of the person or institution from which the specimen were transfered, e.g. the donator of a gift |
YES |
| FromTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
YES |
| FromTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the source of the specimen, e.g. the donating collection of a gift |
YES |
| ToCollectionID |
int |
The ID of the collection to which the specimen were transfered, e.g. the receiver of a gift |
YES |
| ToTransactionPartnerName |
nvarchar (255) |
Name of the person or institution to which the specimen were transfered, e.g. the receiver of a gift |
YES |
| ToTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
YES |
| ToTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the destination of the specimen, e.g. the receiving collection of a gift |
YES |
| ToRecipient |
nvarchar (255) |
The recipient receiving the transaction e.g. if not derived from the link to DiversityAgents |
YES |
| NumberOfUnits |
int |
The number of units which were (initially) included in the transaction |
YES |
| Investigator |
nvarchar (200) |
The investigator for whose study a transacted material was sent |
YES |
| TransactionComment |
nvarchar (MAX) |
Comments on the exchanged material addressed to the transaction partner |
YES |
| BeginDate |
datetime |
Date when the transaction started |
YES |
| AgreedEndDate |
datetime |
End of the transaction period, e.g. if the time for borrowing the specimen is restricted |
YES |
| ActualEndDate |
datetime |
Actual end of the transaction after a prolonation when e.g. the date of return for a loan was prolonged by the owner |
YES |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction not to be published e.g. on a web page |
YES |
| ResponsibleName |
nvarchar (255) |
The person responsible for this transaction |
YES |
| ResponsibleAgentURI |
varchar (255) |
The URI of the person, team or organisation responsible for the data (see e.g. module DiversityAgents) |
YES |
| DateSupplement |
nvarchar (100) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. |
YES |
Depending on:
- CollectionManager
- Transaction
View TransactionPermit
Content of table Transaction restricted to those with TransactionType = permit
| Column |
Data type |
Description |
Nullable |
| TransactionType |
nvarchar (50) |
Type of the transaction, e.g. gift in or out, exchange in or out, purchase in or out |
NO |
| TransactionTitle |
nvarchar (200) |
The title of the transaction as e.g. shown in an user interface |
NO |
| ReportingCategory |
nvarchar (50) |
A group defined for the transaction, e.g. a taxonomic group as used for exchange balancing |
YES |
| AdministratingCollectionID |
int |
ID of the collection which is responsible for the administration of the transaction. |
NO |
| MaterialDescription |
nvarchar (MAX) |
Description of the material of this transaction |
YES |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. |
YES |
| MaterialCollectors |
nvarchar (MAX) |
The collectors of the material |
YES |
| MaterialSource |
nvarchar (500) |
The source of the material within a transaction, e.g. a excavation |
YES |
| FromTransactionPartnerName |
nvarchar (255) |
Name of the person or institution from which the specimen were transfered, e.g. the donator of a gift |
YES |
| FromTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the source of the specimen, e.g. the donating collection of a gift |
YES |
| ToTransactionPartnerName |
nvarchar (255) |
Name of the person or institution to which the specimen were transfered, e.g. the receiver of a gift |
YES |
| ToTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the destination of the specimen, e.g. the receiving collection of a gift |
YES |
| NumberOfUnits |
int |
The number of units which were (initially) included in the transaction |
YES |
| TransactionComment |
nvarchar (MAX) |
Comments on the exchanged material addressed to the transaction partner |
YES |
| Investigator |
nvarchar (200) |
The investigator for whose study a transacted material was sent |
YES |
| BeginDate |
datetime |
Date when the transaction started |
YES |
| AgreedEndDate |
datetime |
End of the transaction period, e.g. if the time for borrowing the specimen is restricted |
YES |
| DateSupplement |
nvarchar (100) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. |
YES |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction not to be published e.g. on a web page |
YES |
| ToRecipient |
nvarchar (255) |
The recipient receiving the transaction e.g. if not derived from the link to DiversityAgents |
YES |
| ResponsibleName |
nvarchar (255) |
The person responsible for this transaction |
YES |
| TransactionID |
int |
Unique ID for the transaction (= primary key) |
NO |
Depending on:
View TransactionRegulation
Content of table Transaction restricted to those with TransactionType = regulation a user has access to
| Column |
Data type |
Description |
Nullable |
| TransactionTitle |
nvarchar (200) |
The title of the transaction as e.g. shown in an user interface |
NO |
| TransactionID |
int |
Unique ID for the transaction (= primary key) |
NO |
Depending on:
- Transaction
- TransactionList
View TransactionRequest
Content of table Transaction with TransactionType loan or request the user has access to as a CollectionManager or the user has created and access to as a CollectionRequester
| Column |
Data type |
Description |
Nullable |
| TransactionID |
int |
Unique ID for the transaction (= primary key) |
NO |
| ParentTransactionID |
int |
The ID of a preceeding transaction of a superior transaction, if transactions are organized in a hierarchy |
YES |
| TransactionType |
nvarchar (50) |
Type of the transaction, e.g. gift in or out, exchange in or out, purchase in or out |
NO |
| TransactionTitle |
nvarchar (200) |
The title of the transaction as e.g. shown in an user interface |
NO |
| ReportingCategory |
nvarchar (50) |
A group defined for the transaction, e.g. a taxonomic group as used for exchange balancing |
YES |
| AdministratingCollectionID |
int |
ID for the collection for which the Manager has the right to administrate the transaction. Corresponds to AdministratingCollectionID in table Transaction. |
NO |
| MaterialDescription |
nvarchar (MAX) |
Description of the material of this transaction |
YES |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. |
YES |
| MaterialCollectors |
nvarchar (MAX) |
The collectors of the material |
YES |
| FromCollectionID |
int |
The ID of the collection from which the specimen were transfered, e.g. the donating collection of a gift |
YES |
| FromTransactionPartnerName |
nvarchar (255) |
Name of the person or institution from which the specimen were transfered, e.g. the donator of a gift |
YES |
| FromTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
YES |
| FromTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the source of the specimen, e.g. the donating collection of a gift |
YES |
| ToCollectionID |
int |
The ID of the collection to which the specimen were transfered, e.g. the receiver of a gift |
YES |
| ToTransactionPartnerName |
nvarchar (255) |
Name of the person or institution to which the specimen were transfered, e.g. the receiver of a gift |
YES |
| ToTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
YES |
| ToTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the destination of the specimen, e.g. the receiving collection of a gift |
YES |
| NumberOfUnits |
smallint |
The number of units which were (initially) included in the transaction |
YES |
| Investigator |
nvarchar (50) |
The investigator for whose study a transacted material was sent |
YES |
| TransactionComment |
nvarchar (MAX) |
Comments on the exchanged material addressed to the transaction partner |
YES |
| BeginDate |
datetime |
Date when the transaction started |
YES |
| AgreedEndDate |
datetime |
End of the transaction period, e.g. if the time for borrowing the specimen is restricted |
YES |
| ActualEndDate |
datetime |
Actual end of the transaction after a prolonation when e.g. the date of return for a loan was prolonged by the owner |
YES |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction not to be published e.g. on a web page |
YES |
| ResponsibleName |
nvarchar (255) |
The person responsible for this transaction |
YES |
| ResponsibleAgentURI |
varchar (255) |
The URI of the person, team or organisation responsible for the data (see e.g. module DiversityAgents) |
YES |
Depending on:
- TransactionForeignRequest
- TransactionUserRequest
View TransactionUserRequest
Content of table Transaction with TransactionType loan or request a CollectionRequester has created and access to
| Column |
Data type |
Description |
Nullable |
| TransactionID |
int |
Unique ID for the transaction (= primary key) |
NO |
| ParentTransactionID |
int |
The ID of a preceeding transaction of a superior transaction, if transactions are organized in a hierarchy |
YES |
| TransactionType |
nvarchar (50) |
Type of the transaction, e.g. gift in or out, exchange in or out, purchase in or out |
NO |
| TransactionTitle |
nvarchar (200) |
The title of the transaction as e.g. shown in an user interface |
NO |
| ReportingCategory |
nvarchar (50) |
A group defined for the transaction, e.g. a taxonomic group as used for exchange balancing |
YES |
| AdministratingCollectionID |
int |
ID for the collection for which the requester has the right to request specimen. Corresponds to AdministratingCollectionID in table Transaction. |
NO |
| MaterialDescription |
nvarchar (MAX) |
Description of the material of this transaction |
YES |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. |
YES |
| MaterialCollectors |
nvarchar (MAX) |
The collectors of the material |
YES |
| FromCollectionID |
int |
The ID of the collection from which the specimen were transfered, e.g. the donating collection of a gift |
YES |
| FromTransactionPartnerName |
nvarchar (255) |
Name of the person or institution from which the specimen were transfered, e.g. the donator of a gift |
YES |
| FromTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
YES |
| FromTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the source of the specimen, e.g. the donating collection of a gift |
YES |
| ToCollectionID |
int |
The ID of the collection to which the specimen were transfered, e.g. the receiver of a gift |
YES |
| ToTransactionPartnerName |
nvarchar (255) |
Name of the person or institution to which the specimen were transfered, e.g. the receiver of a gift |
YES |
| ToTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
YES |
| ToTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the destination of the specimen, e.g. the receiving collection of a gift |
YES |
| NumberOfUnits |
smallint |
The number of units which were (initially) included in the transaction |
YES |
| Investigator |
nvarchar (50) |
The investigator for whose study a transacted material was sent |
YES |
| TransactionComment |
nvarchar (MAX) |
Comments on the exchanged material addressed to the transaction partner |
YES |
| BeginDate |
datetime |
Date when the transaction started |
YES |
| AgreedEndDate |
datetime |
End of the transaction period, e.g. if the time for borrowing the specimen is restricted |
YES |
| ActualEndDate |
datetime |
Actual end of the transaction after a prolonation when e.g. the date of return for a loan was prolonged by the owner |
YES |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction not to be published e.g. on a web page |
YES |
| ResponsibleName |
nvarchar (255) |
The person responsible for this transaction |
YES |
| ResponsibleAgentURI |
varchar (255) |
The URI of the person, team or organisation responsible for the data (see e.g. module DiversityAgents) |
YES |
Depending on:
- CollectionRequester
- Transaction
View UserGroups
List of the groups the users are asigned to
| Column |
Data type |
Description |
Nullable |
| CombinedNameCache |
nvarchar (255) |
The short name of the user, e.g. P. Smith |
NO |
| LoginName |
nvarchar (50) |
A login name which the user uses to access the DivesityWorkbench, Microsoft domains, etc. |
NO |
| UserGroup |
nvarchar (128) |
- |
NO |
Depending on:
View ViewBaseURL
Content of function dbo.BaseURL()
| Column |
Data type |
Description |
Nullable |
| BaseURL |
varchar (255) |
- |
YES |
Depending on:
View ViewCollectionEventImage
Content of table CollectionEventImage converting XML to nvarchar and excluding Log columns
| Column |
Data type |
Description |
Nullable |
| CollectionEventID |
int |
Part of primay key, refers to unique ID for the table CollectionEvent (= foreign key) |
NO |
| URI |
varchar (255) |
The complete URI address of the image. This is only a cached value, if ResourceID is available and referring to the module DiversityResources |
NO |
| ResourceURI |
varchar (255) |
The URI of the resource (e.g. see module DiversityResources) |
YES |
| ImageType |
nvarchar (50) |
Type of the image, e.g. map |
YES |
| Notes |
nvarchar (MAX) |
Notes to this image concerning the CollectionEvent |
YES |
| Description |
nvarchar (MAX) |
Description of the image |
YES |
| Title |
nvarchar (500) |
Title of the resource |
YES |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
| CreatorAgent |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
| CreatorAgentURI |
varchar (255) |
Link to the module DiversityAgents |
YES |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e. g. Creative Common Licenses |
YES |
| InternalNotes |
nvarchar (500) |
Internal notes which should not be published e.g. on websites |
YES |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
| LicenseHolderAgentURI |
nvarchar (500) |
The link to a module containing futher information on the person or institution holding the license |
YES |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
Depending on:
View ViewCollectionEventSeriesImage
Content of table CollectionEventSeriesImage converting XML to nvarchar and excluding Log columns
| Column |
Data type |
Description |
Nullable |
| SeriesID |
int |
Unique ID for the table CollectionEventSeries (= foreign key and part of primary key) |
NO |
| URI |
varchar (255) |
The complete URI address of the image. This is only a cached value, if ResourceID is available and referring to the module DiversityResources |
NO |
| ResourceURI |
varchar (255) |
The URI of the resource (e.g. see module DiversityResources) |
YES |
| ImageType |
nvarchar (50) |
Type of the image, e.g. map |
YES |
| Notes |
nvarchar (MAX) |
Notes to this image of the collection site |
YES |
| Description |
nvarchar (MAX) |
Description of the image |
YES |
| Title |
nvarchar (500) |
Title of the resource |
YES |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
| CreatorAgent |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
| CreatorAgentURI |
varchar (255) |
Link to the module DiversityAgents |
YES |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e.g. Creative Common licenses |
YES |
| InternalNotes |
nvarchar (500) |
Internal notes which should not be published e.g. on websites |
YES |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
| LicenseHolderAgentURI |
nvarchar (500) |
The link to a module containing futher information on the person or institution holding the license |
YES |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
Depending on:
- CollectionEventSeriesImage
View ViewCollectionImage
Content of table Collection converting XML to nvarchar and excluding Log columns
| Column |
Data type |
Description |
Nullable |
| CollectionID |
int |
Refers to the ID of Collection (= foreign key and part of primary key) |
NO |
| URI |
varchar (255) |
The complete URI address of the image. |
NO |
| ImageType |
nvarchar (50) |
Type of the image, e.g. label |
YES |
| Notes |
nvarchar (MAX) |
Notes on the collection image |
YES |
| Description |
nvarchar (MAX) |
Description of the image |
YES |
| Title |
nvarchar (500) |
Title of the resource |
YES |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
| CreatorAgent |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
| CreatorAgentURI |
varchar (255) |
Link to the module DiversityAgents |
YES |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e.g. Creative Common licenses |
YES |
| InternalNotes |
nvarchar (500) |
Internal notes which should not be published e.g. on websites |
YES |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
| LicenseHolderAgentURI |
nvarchar (500) |
The link to a module containing futher information on the person or institution holding the license |
YES |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
Depending on:
View ViewCollectionSpecimenImage
Content of table CollectionSpecimenImage converting XML to nvarchar and excluding Log columns
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
| URI |
varchar (255) |
The complete URI address of the image. This is only a cached value, if ResourceID is available and referring to the module DiversityResources |
NO |
| ResourceURI |
varchar (255) |
The URI of the image, e.g. as stored in the module DiversityResources. |
YES |
| SpecimenPartID |
int |
Optional: If the data set is not related to a part of a specimen, the ID of a related part (= foreign key) |
YES |
| IdentificationUnitID |
int |
If image refers to only one out of several IdentificationUnits for a specimen, refers to the ID of an IdentificationUnit for a collection specimen (= foreign key) |
YES |
| ImageType |
nvarchar (50) |
Type of the image, e.g. photograph |
YES |
| Notes |
nvarchar (MAX) |
Notes on the specimen image |
YES |
| Description |
nvarchar (MAX) |
Description of the image |
YES |
| Title |
nvarchar (500) |
Title of the resource |
YES |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
| CreatorAgent |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
| CreatorAgentURI |
varchar (255) |
Link to the module DiversityAgents |
YES |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e. g. Creative Common Licenses |
YES |
| LicenseURI |
varchar (500) |
The URI of the license for the resource |
YES |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
| LicenseHolderAgentURI |
nvarchar (500) |
The link to a module containing futher information on the person or institution holding the license |
YES |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
| LicenseNotes |
nvarchar (500) |
Notice on license for the resource |
YES |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
| DisplayOrder |
int |
The order in which the images should be shown in a interface |
YES |
| InternalNotes |
nvarchar (500) |
Internal notes which should not be published e.g. on websites |
YES |
Depending on:
View ViewDiversityWorkbenchModule
Content of function dbo.DiversityWorkbenchModule()
| Column |
Data type |
Description |
Nullable |
| DiversityWorkbenchModule |
nvarchar (50) |
- |
YES |
Depending on:
View ViewIdentificationUnitGeoAnalysis
Content of table IdentificationUnitGeoAnalysis converting geography and geometry to nvarchar and excluding Log columns
| Column |
Data type |
Description |
Nullable |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
| IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
NO |
| AnalysisDate |
datetime |
The date of the analysis |
NO |
| Geography |
nvarchar (MAX) |
The geography where the organism resp. object was located according to WGS84, e.g. a point (latitide, longitude and altitude) |
YES |
| Geometry |
nvarchar (MAX) |
The geometry of the place the organism resp. object was observed, e.g. an area |
YES |
| ResponsibleName |
nvarchar (255) |
Name of the person or institution responsible for the determination |
YES |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
YES |
| Notes |
nvarchar (MAX) |
Notes on this analysis |
YES |
Depending on:
- IdentificationUnitGeoAnalysis
Diversity Collection
FUNCTIONS and PROCEDURES
The following objects are not included:
- System objects
- Objects marked as obsolete
- Previous versions of objects
FUNCTIONS
Function AgentOneString
Returns a result set that contains a string with all Collectors of a Specimen
| Parameter |
DataType |
Description |
| @CollectionSpecimenID |
int |
Refers to ID of CollectionEvent (= foreign key and part of primary key) |
| Column |
DataType |
Description |
| CollectionSpecimenID |
int |
Refers to ID of CollectionEvent (= foreign key and part of primary key) |
| CollectorsNameString |
nvarchar (1000) |
- |
| CollectorsNameStringNumber |
nchar (1500) |
- |
Depending on:
Function AnalysisChildNodes
Content of table Analysis containing all children for a given AnalysisID defined via the relation in column AnalysisParentID
| Parameter |
DataType |
Description |
| @ID |
int |
The AnalysisID for which the children should be returned |
| Column |
DataType |
Description |
| AnalysisID |
int |
ID of the analysis (primary key) |
| AnalysisParentID |
int |
Analysis ID of the parent analysis, if it belongs to a certain type documented in this table |
| DisplayText |
nvarchar (50) |
Name of the analysis as e.g. shown in user interface |
| Description |
nvarchar (500) |
Description of the analysis |
| MeasurementUnit |
nvarchar (50) |
The measurement unit used for the analysis, e.g. mm, µmol, kg |
| Notes |
nvarchar (1000) |
Notes on this analysis |
| AnalysisURI |
varchar (255) |
URI referring to an external documentation of the analysis |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries |
| RowGUID |
uniqueidentifier |
- |
Depending on:
Function AnalysisHierarchyAll
Content of table Analysis including the hierarchy separated via “|” defined via the relation in column AnalysisParentID
| Column |
DataType |
Description |
| AnalysisID |
int |
ID of the analysis (primary key) |
| AnalysisParentID |
int |
Analysis ID of the parent analysis, if it belongs to a certain type documented in this table |
| DisplayText |
nvarchar (50) |
Name of the analysis as e.g. shown in user interface |
| Description |
nvarchar (MAX) |
Description of the analysis |
| MeasurementUnit |
nvarchar (50) |
The measurement unit used for the analysis, e.g. mm, µmol, kg |
| Notes |
nvarchar (MAX) |
Notes on this analysis |
| AnalysisURI |
varchar (255) |
URI referring to an external documentation of the analysis |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries |
| HierarchyDisplayText |
varchar (900) |
- |
Depending on:
Function AnalysisList
Returns the content of table Analysis related to the given project and a taxonomic group
| Parameter |
DataType |
Description |
| @ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
| @TaxonomicGroup |
nvarchar (50) |
Taxonomic group the organism, identified by this unit, belongs to. Groups listed in table CollTaxonomicGroup_Enum (= foreign key) |
| Column |
DataType |
Description |
| AnalysisID |
int |
ID of the analysis (primary key) |
| AnalysisParentID |
int |
Analysis ID of the parent analysis, if it belongs to a certain type documented in this table |
| DisplayText |
nvarchar (50) |
Name of the analysis as e.g. shown in user interface |
| Description |
nvarchar (500) |
Description of the analysis |
| MeasurementUnit |
nvarchar (50) |
The measurement unit used for the analysis, e.g. mm, µmol, kg |
| Notes |
nvarchar (1000) |
Notes on this analysis |
| AnalysisURI |
varchar (255) |
URI referring to an external documentation of the analysis |
| DisplayTextHierarchy |
nvarchar (255) |
- |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries |
Depending on:
- Analysis
- AnalysisChildNodes
- AnalysisTaxonomicGroup
- ProjectAnalysis
Function AnalysisListForUnit
Returns the content of table Analysis available for a IdentificationUnit given by the IdentificationUnitID. The list depends upon the analysis types available for a taxonomic group and the projects available for an analysis
| Parameter |
DataType |
Description |
| @IdentificationUnitID |
int |
ID of the IdentificationUnit (= part of primary key). Usually one of possibly several organisms present on the collected specimen. Example: parasite with hyperparasite on plant leaf = 3 units, |
| Column |
DataType |
Description |
| AnalysisID |
int |
ID of the analysis (primary key) |
| AnalysisParentID |
int |
Analysis ID of the parent analysis, if it belongs to a certain type documented in this table |
| DisplayText |
nvarchar (50) |
Name of the analysis as e.g. shown in user interface |
| Description |
nvarchar (1000) |
Description of the analysis |
| MeasurementUnit |
nvarchar (50) |
The measurement unit used for the analysis, e.g. mm, µmol, kg |
| Notes |
nvarchar (1000) |
Notes on this analysis |
| AnalysisURI |
varchar (255) |
URI referring to an external documentation of the analysis |
| DisplayTextHierarchy |
nvarchar (255) |
- |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries |
Depending on:
- Analysis
- AnalysisTaxonomicGroup
- CollectionProject
- IdentificationUnit
- ProjectAnalysis
Function AnalysisProjectList
Returns content of table Analysis related to the given project
| Parameter |
DataType |
Description |
| @ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
| Column |
DataType |
Description |
| AnalysisID |
int |
ID of the analysis (primary key) |
| AnalysisParentID |
int |
Analysis ID of the parent analysis, if it belongs to a certain type documented in this table |
| DisplayText |
nvarchar (50) |
Name of the analysis as e.g. shown in user interface |
| Description |
nvarchar (500) |
Description of the analysis |
| MeasurementUnit |
nvarchar (50) |
The measurement unit used for the analysis, e.g. mm, µmol, kg |
| Notes |
nvarchar (1000) |
Notes on this analysis |
| AnalysisURI |
varchar (255) |
URI referring to an external documentation of the analysis |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries |
| RowGUID |
uniqueidentifier |
- |
Depending on:
- Analysis
- AnalysisChildNodes
- ProjectAnalysis
Function AnalysisTaxonomicGroupForProject
Returns the content of the table AnalysisTaxonomicGroup used in a project including the whole hierarchy
| Parameter |
DataType |
Description |
| @ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
| Column |
DataType |
Description |
| AnalysisID |
int |
ID of the analysis (primary key) |
| TaxonomicGroup |
nvarchar (50) |
Taxonomic group the organism, identified by this unit, belongs to. Groups listed in table CollTaxonomicGroup_Enum (= foreign key) |
| RowGUID |
uniqueidentifier |
- |
Depending on:
- Analysis
- AnalysisList
- AnalysisTaxonomicGroup
- ProjectAnalysis
Function AverageAltitude
The average altitude for the points of a geography where the z value is given
DataType: float
| Parameter |
DataType |
Description |
| @Geography |
geography (-1) |
The geography for which the average altitude should be extracted |
Function BaseURL
Returns the basic URL for the database
DataType: varchar (255)
Function CollCharacterType_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| Code |
nvarchar (50) |
- |
| Description |
nvarchar (500) |
- |
| Abbreviation |
nvarchar (50) |
- |
Function CollDateCategory_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| Code |
nvarchar (50) |
A text code that uniquely identifies each object in the enumeration (primary key). This value may not be changed, because the application may depend upon it. |
| Description |
nvarchar (500) |
Description of enumerated object, displayed in the user interface |
| Abbreviation |
nvarchar (50) |
- |
Function CollectionChildNodes
Returns a result set that lists all the items within a hierarchy starting at the topmost item related to the given item.
| Parameter |
DataType |
Description |
| @ID |
int |
ID of the collection (= CollectionID, PK) |
| Column |
DataType |
Description |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
| CollectionName |
nvarchar (255) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
| CollectionAcronym |
nvarchar (10) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
| AdministrativeContactAgentURI |
nvarchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
| Description |
nvarchar (MAX) |
A short description of the collection |
| Location |
nvarchar (255) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
| LocationPlan |
varchar (500) |
URI or file name including path of the floor plan of the collection |
| LocationPlanWidth |
float |
Width of location plan in meter for calculation of size by provided geometry |
| LocationPlanDate |
datetime |
The date when the plan for the collection has been created |
| LocationHeight |
float |
Height from ground level, e.g. for the position of sensors |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
| Type |
nvarchar (50) |
Type of the collection, e.g. cupboard, drawer etc. |
| LocationParentID |
int |
If the hierarchy of the location does not match the logical hierarchy, the ID of the parent location |
Depending on:
Function CollectionEventSeriesHierarchy
Returns a table that lists all the Series related to the given Series.
| Parameter |
DataType |
Description |
| @SeriesID |
int |
Primary key. The ID for this series of collection events (= primary key) |
| Column |
DataType |
Description |
| SeriesID |
int |
Primary key. The ID for this series of collection events (= primary key) |
| SeriesParentID |
int |
The ID of the superior series of collection events |
| DateStart |
datetime |
Point in time when the series of collection events started |
| DateEnd |
datetime |
Point in time when the series of collection events ended |
| DateSupplement |
nvarchar (100) |
Verbal or additional collection date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. The end date, if the collection event series comprises a period. The time of the event, if necessary. |
| SeriesCode |
nvarchar (50) |
The user defined code for a series of collection events |
| Description |
nvarchar (500) |
The description of the series of collection events as it will be printed on e.g. the label |
| Notes |
nvarchar (500) |
Notes on this series of collection events |
| Geography |
geography (MAX) |
The geography of the series of collection events |
Depending on:
- CollectionEventSeries
- EventSeriesChildNodes
- EventSeriesTopID
Function CollectionHierarchy
Returns a table that lists all the analysis items related to the given analysis.
| Parameter |
DataType |
Description |
| @CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| Column |
DataType |
Description |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
| CollectionName |
nvarchar (255) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
| CollectionAcronym |
nvarchar (10) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
| AdministrativeContactAgentURI |
nvarchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
| Description |
nvarchar (MAX) |
A short description of the collection |
| Location |
nvarchar (255) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
| LocationPlan |
varchar (500) |
URI or file name including path of the floor plan of the collection |
| LocationPlanWidth |
float |
Width of location plan in meter for calculation of size by provided geometry |
| LocationPlanDate |
datetime |
The date when the plan for the collection has been created |
| LocationHeight |
float |
Height from ground level, e.g. for the position of sensors |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
| Type |
nvarchar (50) |
Type of the collection, e.g. cupboard, drawer etc. |
| LocationParentID |
int |
If the hierarchy of the location does not match the logical hierarchy, the ID of the parent location |
Depending on:
- Collection
- CollectionChildNodes
Function CollectionHierarchyAll
Returns a table that lists all the collections including a display text with the whole hierarchy
| Column |
DataType |
Description |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
| CollectionName |
nvarchar (500) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
| CollectionAcronym |
nvarchar (50) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
| AdministrativeContactAgentURI |
nvarchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
| Description |
nvarchar (MAX) |
A short description of the collection |
| Location |
nvarchar (255) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
| LocationPlan |
varchar (500) |
URI or file name including path of the floor plan of the collection |
| LocationPlanWidth |
float |
Width of location plan in meter for calculation of size by provided geometry |
| LocationPlanDate |
datetime |
The date when the plan for the collection has been created |
| LocationHeight |
float |
Height from ground level, e.g. for the position of sensors |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
| DisplayOrder |
varchar (255) |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
| DisplayText |
varchar (900) |
- |
| Type |
nvarchar (50) |
Type of the collection, e.g. cupboard, drawer etc. |
| LocationParentID |
int |
If the hierarchy of the location does not match the logical hierarchy, the ID of the parent location |
Depending on:
- Collection
- UserCollectionList
Function CollectionHierarchyMulti
Returns a table that lists all the collections related to the given collection in the list.
| Parameter |
DataType |
Description |
| @CollectionIDs |
varchar (255) |
Comma separated list of IDs of the collections that should be included. |
| Column |
DataType |
Description |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
| CollectionName |
nvarchar (50) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
| CollectionAcronym |
nvarchar (50) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
| AdministrativeContactAgentURI |
nvarchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
| Description |
nvarchar (500) |
A short description of the collection |
| Location |
nvarchar (255) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
| LocationPlan |
varchar (500) |
URI or file name including path of the floor plan of the collection |
| LocationPlanWidth |
float |
Width of location plan in meter for calculation of size by provided geometry |
| LocationPlanDate |
datetime |
The date when the plan for the collection has been created |
| LocationHeight |
float |
Height from ground level, e.g. for the position of sensors |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
| DisplayOrder |
varchar (255) |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
| Type |
nvarchar (50) |
Type of the collection, e.g. cupboard, drawer etc. |
| LocationParentID |
int |
If the hierarchy of the location does not match the logical hierarchy, the ID of the parent location |
Depending on:
Function CollectionHierarchySuperior
Returns a table that lists the given and all the items superior to the given collection
| Parameter |
DataType |
Description |
| @CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| Column |
DataType |
Description |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
| CollectionName |
nvarchar (255) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
| CollectionAcronym |
nvarchar (10) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
| AdministrativeContactAgentURI |
nvarchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
| Description |
nvarchar (MAX) |
A short description of the collection |
| Location |
nvarchar (255) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
| LocationPlan |
varchar (500) |
URI or file name including path of the floor plan of the collection |
| LocationPlanWidth |
float |
Width of location plan in meter for calculation of size by provided geometry |
| LocationPlanDate |
datetime |
The date when the plan for the collection has been created |
| LocationHeight |
float |
Height from ground level, e.g. for the position of sensors |
| LocationGeometry |
geometry (MAX) |
Geometry of the collection within the floor plan |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
| Type |
nvarchar (50) |
Type of the collection, e.g. cupboard, drawer etc. |
| LocationParentID |
int |
If the hierarchy of the location does not match the logical hierarchy, the ID of the parent location |
Depending on:
Function CollectionLocation
Returns a table that lists all the collection items related to the given collection
| Parameter |
DataType |
Description |
| @CollectionID |
int |
The ID of the collection for which the hierarchy should be listed |
| Column |
DataType |
Description |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
| CollectionName |
nvarchar (255) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
| CollectionAcronym |
nvarchar (10) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
| AdministrativeContactAgentURI |
nvarchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
| Description |
nvarchar (MAX) |
A short description of the collection |
| Location |
nvarchar (255) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
| LocationPlan |
varchar (500) |
URI or file name including path of the floor plan of the collection |
| LocationPlanWidth |
float |
Width of location plan in meter for calculation of size by provided geometry |
| LocationPlanDate |
datetime |
The date when the plan for the collection has been created |
| LocationHeight |
float |
Height from ground level, e.g. for the position of sensors |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
| Type |
nvarchar (50) |
Type of the collection, e.g. cupboard, drawer etc. |
| LocationParentID |
int |
If the hierarchy of the location does not match the logical hierarchy, the ID of the parent location |
Depending on:
- Collection
- CollectionLocationChildNodes
Function CollectionLocationAll
Returns a table that lists all the collections including a display text with the whole hierarchy
| Column |
DataType |
Description |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
| CollectionName |
nvarchar (500) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
| CollectionAcronym |
nvarchar (50) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
| AdministrativeContactAgentURI |
nvarchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
| Description |
nvarchar (MAX) |
A short description of the collection |
| Location |
nvarchar (255) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
| LocationPlan |
varchar (500) |
URI or file name including path of the floor plan of the collection |
| LocationPlanWidth |
float |
Width of location plan in meter for calculation of size by provided geometry |
| LocationPlanDate |
datetime |
The date when the plan for the collection has been created |
| LocationHeight |
float |
Height from ground level, e.g. for the position of sensors |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
| DisplayOrder |
varchar (255) |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
| DisplayText |
varchar (900) |
Hierarchy of the collection with acronym if present, otherwise name |
| Type |
nvarchar (50) |
Type of the collection, e.g. cupboard, drawer etc. |
| LocationParentID |
int |
If the hierarchy of the location does not match the logical hierarchy, the ID of the parent location |
Depending on:
- Collection
- UserCollectionList
Function CollectionLocationChildNodes
Returns a result set that lists all the items within a hierarchy starting at the topmost item related to the given item
| Parameter |
DataType |
Description |
| @ID |
int |
The ID of the collection for which the hierarchy should be listed |
| Column |
DataType |
Description |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
| CollectionName |
nvarchar (255) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
| CollectionAcronym |
nvarchar (10) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
| AdministrativeContactAgentURI |
nvarchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
| Description |
nvarchar (MAX) |
A short description of the collection |
| Location |
nvarchar (255) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
| LocationPlan |
varchar (500) |
URI or file name including path of the floor plan of the collection |
| LocationPlanWidth |
float |
Width of location plan in meter for calculation of size by provided geometry |
| LocationPlanDate |
datetime |
The date when the plan for the collection has been created |
| LocationHeight |
float |
Height from ground level, e.g. for the position of sensors |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
| Type |
nvarchar (50) |
Type of the collection, e.g. cupboard, drawer etc. |
| LocationParentID |
int |
If the hierarchy of the location does not match the logical hierarchy, the ID of the parent location |
Depending on:
Function CollectionLocationMulti
List of collections according to the given CollectionIDs
| Parameter |
DataType |
Description |
| @CollectionIDs |
varchar (4000) |
Space separated list of CollectionIDs with a maximal length of 4000 |
| Column |
DataType |
Description |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
| CollectionName |
nvarchar (50) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
| CollectionAcronym |
nvarchar (50) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
| AdministrativeContactAgentURI |
nvarchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
| Description |
nvarchar (500) |
A short description of the collection |
| Location |
nvarchar (255) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
| LocationPlan |
varchar (500) |
URI or file name including path of the floor plan of the collection |
| LocationPlanWidth |
float |
Width of location plan in meter for calculation of size by provided geometry |
| LocationPlanDate |
datetime |
The date when the plan for the collection has been created |
| LocationHeight |
float |
Height from ground level, e.g. for the position of sensors |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
| DisplayOrder |
varchar (255) |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
| Type |
nvarchar (50) |
Type of the collection, e.g. cupboard, drawer etc. |
| LocationParentID |
int |
If the hierarchy of the location does not match the logical hierarchy, the ID of the parent location |
Depending on:
Function CollectionLocationSuperior
Returns a table that lists the given and all the items superior to the given collection
| Parameter |
DataType |
Description |
| @CollectionID |
int |
The ID of the collection for which the hierarchy should be listed |
| Column |
DataType |
Description |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
| CollectionName |
nvarchar (255) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
| CollectionAcronym |
nvarchar (10) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
| AdministrativeContactAgentURI |
nvarchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
| Description |
nvarchar (MAX) |
A short description of the collection |
| Location |
nvarchar (255) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
| LocationPlan |
varchar (500) |
URI or file name including path of the floor plan of the collection |
| LocationPlanWidth |
float |
Width of location plan in meter for calculation of size by provided geometry |
| LocationPlanDate |
datetime |
The date when the plan for the collection has been created |
| LocationHeight |
float |
Height from ground level, e.g. for the position of sensors |
| LocationGeometry |
geometry (MAX) |
Geometry of the collection within the floor plan |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
| Type |
nvarchar (50) |
Type of the collection, e.g. cupboard, drawer etc. |
| LocationParentID |
int |
If the hierarchy of the location does not match the logical hierarchy, the ID of the parent location |
Depending on:
Function CollectionSpecimenCoordinateList
| Parameter |
DataType |
Description |
| @CollectionSpecimenIDList |
varchar (8000) |
- |
| Column |
DataType |
Description |
| CollectionSpecimenID |
int |
Unique ID for the table CollectionSpecimen (primary key) |
| Latitude |
float |
- |
| Longitude |
float |
- |
Depending on:
- CollectionSpecimenCoordinates
Function CollectionSpecimenCoordinates
| Parameter |
DataType |
Description |
| @CollectionSpecimenID |
int |
Unique ID for the table CollectionSpecimen (primary key) |
| Column |
DataType |
Description |
| CollectionSpecimenID |
int |
Unique ID for the table CollectionSpecimen (primary key) |
| Latitude |
float |
- |
| Longitude |
float |
- |
| LocationAccuracy |
nvarchar (50) |
The accuracy of the determination of this locality |
| DistanceToLocation |
nvarchar (50) |
Distance from the specified place to the real location of the collection site (m) |
| DirectionToLocation |
nvarchar (50) |
Direction from the specified place to the real location of the collection site (Degrees rel. to north) |
Depending on:
- CollectionEventLocalisation
- CollectionSpecimen
Function CollectionSpecimenRelationInversList
Listing all internal relations in reverse perspective
| Column |
DataType |
Description |
| CollectionSpecimenID |
int |
Unique ID for the table CollectionSpecimen (primary key) |
| RelatedSpecimenCollectionSpecimenID |
int |
The CollectionSpecimenID of the dataset that refers to the current dataset |
| RelatedSpecimenAccessionNumber |
nvarchar (50) |
The AccessionNumber of the dataset that refers to the current dataset |
| RelationType |
nvarchar (50) |
Type of the relation between the specimen (= foreign key, see table CollRelationType_Enum) |
| RelatedSpecimenCollectionID |
int |
ID of the Collection as stored in table Collection (= foreign key, see table Collection) |
| Notes |
nvarchar (MAX) |
Notes on the relation to the specimen |
Depending on:
- BaseURL
- CollectionSpecimen
- CollectionSpecimenRelation
Function CollectionTaskChildNodes
Returns a result set that lists all children of a collection task within a hierarchy
| Parameter |
DataType |
Description |
| @ID |
int |
The CollectionTaskID to which the inferior datasets are link to via column CollectionTaskParentID |
| Column |
DataType |
Description |
| CollectionTaskID |
int |
PK of the table |
| CollectionTaskParentID |
int |
Relation to PK for hierarchy within the data |
| CollectionID |
int |
Relation to table Collection. Every ColletionTask needs a relation to a collection |
| TaskID |
int |
Relation to table Task where details for the collection task are defined |
| DisplayOrder |
int |
Display order e.g. in a report. Data with value 0 will not be included |
| DisplayText |
nvarchar (400) |
The display text of the module related data as shown e.g. in a user interface |
| ModuleUri |
varchar (500) |
The URL of module related data |
| CollectionSpecimenID |
int |
Refers to the CollectionSpecimenID of CollectionSpecimenPart (= foreign key and part of primary key) |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen the task is related to. |
| TransactionID |
int |
ID of a transaction. Related to PK of table Transaction |
| TaskStart |
datetime |
The start date and or time of the collection tasks. The type is defined in table Task |
| TaskEnd |
datetime |
The end date and or time of the collection tasks. The type is defined in table Task |
| Result |
nvarchar (400) |
A text result either taken from a list or entered. The type is defined in table Task |
| URI |
varchar (500) |
The URI of the collection tasks. The type is defined in table Task |
| NumberValue |
real |
The numeric value of the collection tasks. The type is defined in table Task |
| BoolValue |
bit |
The boolean of the collection tasks. The type is defined in table Task |
| MetricDescription |
nvarchar (500) |
Description of the metric e.g. imported from a time series database like Prometheus |
| MetricSource |
varchar (4000) |
The source of the metric e.g. the PromQL statement for the import from a timeseries database Prometheus |
| MetricUnit |
nvarchar (50) |
The unit of the metric, e.g. °C for temperature |
| ResponsibleAgent |
nvarchar (500) |
The name of the responsible person or institution |
| ResponsibleAgentURI |
varchar (500) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
| Description |
nvarchar (MAX) |
The description of the collection tasks. The type is defined in table Task |
| Notes |
nvarchar (MAX) |
The notes of the collection tasks. The type is defined in table Task |
Depending on:
Function CollectionTaskCollectionHierarchyAll
Returns a result set that lists all the collection tasks including a display text with the related collection as leading part
| Column |
DataType |
Description |
| CollectionTaskID |
int |
PK of the table |
| CollectionTaskParentID |
int |
Relation to PK for hierarchy within the data |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| TaskID |
int |
Relation to table Task where details for the collection task are defined |
| DisplayText |
nvarchar (400) |
The display text of the module related data as shown e.g. in a user interface |
| DisplayOrder |
int |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
| ModuleUri |
varchar (500) |
The URL of module related data |
| CollectionSpecimenID |
int |
Refers to the CollectionSpecimenID of CollectionSpecimenPart (= foreign key and part of primary key) |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen the task is related to. |
| TransactionID |
int |
The ID of the transaction to which the collection task is linked |
| TaskStart |
datetime |
The start date and or time of the collection tasks. The type is defined in table Task |
| TaskEnd |
datetime |
The end date and or time of the collection tasks. The type is defined in table Task |
| Result |
nvarchar (400) |
A text result either taken from a list or entered. The type is defined in table Task |
| URI |
varchar (500) |
The URI of the collection tasks. The type is defined in table Task |
| NumberValue |
real |
The numeric value of the collection tasks. The type is defined in table Task |
| BoolValue |
bit |
The boolean of the collection tasks. The type is defined in table Task |
| MetricDescription |
nvarchar (500) |
Description of the metric e.g. imported from a time series database like Prometheus |
| MetricSource |
varchar (4000) |
The source of the metric e.g. the PromQL statement for the import from a timeseries database Prometheus |
| MetricUnit |
nvarchar (50) |
The unit of the metric, e.g. °C for temperature |
| ResponsibleAgent |
nvarchar (500) |
The name of the responsible person or institution |
| ResponsibleAgentURI |
varchar (500) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
| Description |
nvarchar (MAX) |
A short description of the collection |
| Notes |
nvarchar (MAX) |
The notes of the collection tasks. The type is defined in table Task |
| HierarchyDisplayText |
nvarchar (900) |
Display text including the hierarchy of the collection separated by the string defined in function TaskCollectionHierarchySeparator followed by the hierarchy of the collection task separated by the string defined in function TaskHierarchySeparator followed by optional information about start, end and result of the collection task |
Depending on:
- CollectionHierarchyAll
- CollectionTask
- Task
- TaskCollectionHierarchySeparator
- TaskHierarchySeparator
Function CollectionTaskHierarchy
Returns a result set that lists all the collection tasks within a hierarchy starting at the topmost collection task related to the given collection task
| Parameter |
DataType |
Description |
| @ID |
int |
The CollectionTaskID for which the hierarchy should be retrieved |
| Column |
DataType |
Description |
| CollectionTaskID |
int |
PK of the table |
| CollectionTaskParentID |
int |
Relation to PK for hierarchy within the data |
| CollectionID |
int |
Relation to table Collection. Every ColletionTask needs a relation to a collection |
| TaskID |
int |
Relation to table Task where details for the collection task are defined |
| DisplayOrder |
int |
Display order e.g. in a report. Data with value 0 will not be included |
| DisplayText |
nvarchar (400) |
The display text of the module related data as shown e.g. in a user interface |
| ModuleUri |
varchar (500) |
The URL of module related data |
| CollectionSpecimenID |
int |
Refers to the CollectionSpecimenID of CollectionSpecimenPart (= foreign key and part of primary key) |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen the task is related to. |
| TransactionID |
int |
The ID of the transaction to which the collection task is linked |
| TaskStart |
datetime |
The start date and or time of the collection tasks. The type is defined in table Task |
| TaskEnd |
datetime |
The end date and or time of the collection tasks. The type is defined in table Task |
| Result |
nvarchar (400) |
A text result either taken from a list or entered. The type is defined in table Task |
| URI |
varchar (500) |
The URI of the collection tasks. The type is defined in table Task |
| NumberValue |
real |
The numeric value of the collection tasks. The type is defined in table Task |
| BoolValue |
bit |
The boolean of the collection tasks. The type is defined in table Task |
| MetricDescription |
nvarchar (500) |
Description of the metric e.g. imported from a time series database like Prometheus |
| MetricSource |
varchar (4000) |
The source of the metric e.g. the PromQL statement for the import from a timeseries database Prometheus |
| MetricUnit |
nvarchar (50) |
The unit of the metric, e.g. °C for temperature |
| ResponsibleAgent |
nvarchar (500) |
The name of the responsible person or institution |
| ResponsibleAgentURI |
varchar (500) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
| Description |
nvarchar (MAX) |
The description of the collection tasks. The type is defined in table Task |
| Notes |
nvarchar (MAX) |
The notes of the collection tasks. The type is defined in table Task |
Depending on:
- CollectionTask
- CollectionTaskChildNodes
Function CollectionTaskHierarchyAll
All CollectionTasks including a column displaying the hierarchy of the CollectionTask
| Column |
DataType |
Description |
| CollectionTaskID |
int |
PK of the table |
| CollectionTaskParentID |
int |
Relation to PK for hierarchy within the data |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| TaskID |
int |
Relation to table Task where details for the collection task are defined |
| DisplayOrder |
int |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
| Task |
nvarchar (400) |
Display text of the task the collection task is linked to |
| DisplayText |
nvarchar (400) |
The display text of the module related data as shown e.g. in a user interface |
| ModuleUri |
varchar (500) |
The URL of module related data |
| CollectionSpecimenID |
int |
Refers to the CollectionSpecimenID of CollectionSpecimenPart (= foreign key and part of primary key) |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen the task is related to. |
| TransactionID |
int |
The ID of the transaction to which the collection task is linked |
| TaskStart |
datetime |
The start date and or time of the collection tasks. The type is defined in table Task |
| TaskEnd |
datetime |
The end date and or time of the collection tasks. The type is defined in table Task |
| Result |
nvarchar (400) |
A text result either taken from a list or entered. The type is defined in table Task |
| URI |
varchar (500) |
The URI of the collection tasks. The type is defined in table Task |
| NumberValue |
real |
The numeric value of the collection tasks. The type is defined in table Task |
| BoolValue |
bit |
The boolean of the collection tasks. The type is defined in table Task |
| MetricDescription |
nvarchar (500) |
Description of the metric e.g. imported from a time series database like Prometheus |
| MetricSource |
varchar (4000) |
The source of the metric e.g. the PromQL statement for the import from a timeseries database Prometheus |
| MetricUnit |
nvarchar (50) |
The unit of the metric, e.g. °C for temperature |
| ResponsibleAgent |
nvarchar (500) |
The name of the responsible person or institution |
| ResponsibleAgentURI |
varchar (500) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
| Description |
nvarchar (MAX) |
A short description of the collection |
| Notes |
nvarchar (MAX) |
The notes of the collection tasks. The type is defined in table Task |
| HierarchyDisplayText |
nvarchar (4000) |
Display text including the hierarchy of the collection task separated by the string defined in function TaskHierarchySeparator followed by the hierarchy of the collection separated by the string defined in function TaskCollectionHierarchySeparator |
| CollectionHierarchyDisplayText |
nvarchar (4000) |
Display text including the hierarchy of the collection separated by the string defined in function TaskCollectionHierarchySeparator followed by the hierarchy of the collection task separated by the string defined in function TaskHierarchySeparator |
| TaskHierarchyDisplayText |
nvarchar (4000) |
Deprecated |
| TaskDisplayText |
nvarchar (4000) |
Display text of the collection task based on diverse content in tables Task and CollectionTask |
Depending on:
- CollectionHierarchyAll
- CollectionTask
- Task
- TaskCollectionHierarchySeparator
- TaskHierarchySeparator
Function CollectionTaskParentNodes
Parent CollectionTasks for a given CollectionTask in the hierarchy
| Parameter |
DataType |
Description |
| @ID |
int |
The CollectionTaskID for which the parent hierarchy should be retrieved |
| Column |
DataType |
Description |
| CollectionTaskID |
int |
PK of the table |
| CollectionTaskParentID |
int |
Relation to PK for hierarchy within the data |
| CollectionID |
int |
Relation to table Collection. Every ColletionTask needs a relation to a collection |
| TaskID |
int |
Relation to table Task where details for the collection task are defined |
| DisplayOrder |
int |
Display order e.g. in a report. Data with value 0 will not be included |
| DisplayText |
nvarchar (400) |
The display text of the module related data as shown e.g. in a user interface |
| ModuleUri |
varchar (500) |
The URL of module related data |
| CollectionSpecimenID |
int |
Refers to the CollectionSpecimenID of CollectionSpecimenPart (= foreign key and part of primary key) |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen the task is related to. |
| TransactionID |
int |
ID of a transaction. Related to PK of table Transaction |
| TaskStart |
datetime |
The start date and or time of the collection tasks. The type is defined in table Task |
| TaskEnd |
datetime |
The end date and or time of the collection tasks. The type is defined in table Task |
| Result |
nvarchar (400) |
A text result either taken from a list or entered. The type is defined in table Task |
| URI |
varchar (500) |
The URI of the collection tasks. The type is defined in table Task |
| NumberValue |
real |
The numeric value of the collection tasks. The type is defined in table Task |
| BoolValue |
bit |
The boolean of the collection tasks. The type is defined in table Task |
| MetricDescription |
nvarchar (500) |
Description of the metric e.g. imported from a time series database like Prometheus |
| MetricSource |
varchar (4000) |
The source of the metric e.g. the PromQL statement for the import from a timeseries database Prometheus |
| MetricUnit |
nvarchar (50) |
The unit of the metric, e.g. °C for temperature |
| ResponsibleAgent |
nvarchar (500) |
The name of the responsible person or institution |
| ResponsibleAgentURI |
varchar (500) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
| Description |
nvarchar (MAX) |
The description of the collection tasks. The type is defined in table Task |
| Notes |
nvarchar (MAX) |
The notes of the collection tasks. The type is defined in table Task |
Depending on:
Function CollEventDateCategory_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| Code |
nvarchar (50) |
A text code which uniquely identifies each object in the enumeration (primary key). This value may not be changed, because the application may depend upon it. |
| Description |
nvarchar (500) |
Description of enumerated object, displayed in the user interface |
| Abbreviation |
nvarchar (50) |
- |
Function CollEventImageType_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| Code |
nvarchar (50) |
A text code which uniquely identifies each object in the enumeration (primary key). This value may not be changed, because the application may depend upon it. |
| Description |
nvarchar (500) |
Description of enumerated object, displayed in the user interface |
| Abbreviation |
nvarchar (50) |
- |
Function CollExchangeType_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| Code |
nvarchar (50) |
- |
| Description |
nvarchar (500) |
- |
| Abbreviation |
nvarchar (50) |
- |
Function CollIdentificationCategory_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| Code |
nvarchar (50) |
A text code which uniquely identifies each object in the enumeration (primary key). This value may not be changed, because the application may depend upon it. |
| Description |
nvarchar (500) |
Description of enumerated object, displayed in the user interface |
| Abbreviation |
nvarchar (50) |
- |
Function CollIdentificationDateCategory_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| Code |
nvarchar (50) |
A text code which uniquely identifies each object in the enumeration (primary key). This value may not be changed, because the application may depend upon it. |
| Description |
nvarchar (500) |
Description of enumerated object, displayed in the user interface |
| Abbreviation |
nvarchar (50) |
- |
Function CollIdentificationQualifier_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| Code |
nvarchar (50) |
A text code which uniquely identifies each object in the enumeration (primary key). This value may not be changed, because the application may depend upon it. |
| Description |
nvarchar (500) |
Description of enumerated object, displayed in the user interface |
| Abbreviation |
nvarchar (50) |
- |
Function CollLabelType_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| Code |
nvarchar (50) |
A text code which uniquely identifies each object in the enumeration (primary key). This value may not be changed, because the application may depend upon it. |
| Description |
nvarchar (500) |
Description of enumerated object, displayed in the user interface |
| Abbreviation |
nvarchar (50) |
- |
Function CollMaterialCategory_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| Code |
nvarchar (50) |
A text code which uniquely identifies each object in the enumeration (primary key). This value may not be changed, because the application may depend upon it. |
| Description |
nvarchar (500) |
Description of enumerated object, displayed in the user interface |
| Abbreviation |
nvarchar (50) |
- |
Function CollSpecimenImageType_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| Code |
nvarchar (50) |
A text code which uniquely identifies each object in the enumeration (primary key). This value may not be changed, because the application may depend upon it. |
| Description |
nvarchar (500) |
Description of enumerated object, displayed in the user interface |
| Abbreviation |
nvarchar (50) |
- |
Function CollTranscriptionState_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| Code |
nvarchar (50) |
- |
| Description |
nvarchar (500) |
- |
| Abbreviation |
nvarchar (50) |
- |
Function CollTypeStatus_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| Code |
nvarchar (50) |
A text code which uniquely identifies each object in the enumeration (primary key). This value may not be changed because the application may depend upon it. |
| Description |
nvarchar (500) |
Description of enumerated object, displayed in the user interface |
| Abbreviation |
nvarchar (50) |
- |
Function ColTaxonomicGroup_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| Code |
nvarchar (50) |
A text code which uniquely identifies each object in the enumeration (primary key). This value may not be changed, because the application may depend upon it. |
| Description |
nvarchar (500) |
Description of enumerated object, displayed in the user interface |
| Abbreviation |
nvarchar (50) |
- |
Function CuratorCollectionHierarchyList
| Column |
DataType |
Description |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
| CollectionName |
nvarchar (50) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
| CollectionAcronym |
nvarchar (50) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
| AdministrativeContactAgentURI |
nvarchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
| Description |
nvarchar (500) |
A short description of the collection |
| Location |
nvarchar (1000) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
| DisplayOrder |
varchar (255) |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
Depending on:
- Collection
- CollectionHierarchy
- CuratorCollectionList
Function CurrentUser
Returns the information for the current user
| Column |
DataType |
Description |
| LoginName |
nvarchar (50) |
A login name which the user uses to access the DivesityWorkbench, Microsoft domains, etc. |
| CombinedNameCache |
nvarchar (255) |
The short name of the user, e.g. P. Smith |
| UserURI |
varchar (255) |
Deprecated. URI of a user in a remote module, e.g. refering to UserInfo.UserID in database DiversityUsers |
| AgentURI |
varchar (255) |
URI of a agent in the module DiversityAgents |
| UserName |
nvarchar (255) |
- |
Depending on:
Function CurrentUserName
retrieval of the name of the current user
DataType: nvarchar (4000)
Depending on:
Function DefaultProjectID
Returns the ID of the current project used by the user
DataType: int
Depending on:
Function DiversityCollectionCacheDatabaseName
returns the name of the cache database containing the exported data for e.g. publication on the web
DataType: nvarchar (50)
Function DiversityWorkbenchModule
Name of the module
DataType: nvarchar (50)
Returns the information about an entity
| Parameter |
DataType |
Description |
| @Entity |
varchar (500) |
The name of the entity, e.g. Table.Column.Content within the database or a unique string for e.g. a message within the DiversityWorkbench e.g. “DiversityWorkbench.Message.Connection.NoAccess”, PK |
| @Language |
nvarchar (50) |
The language to which the information should be restricted |
| @Context |
nvarchar (50) |
The context to which the information should be restricted |
| Column |
DataType |
Description |
| Entity |
varchar (500) |
The name of the entity, e.g. Table.Column.Content within the database or a unique string for e.g. a message within the DiversityWorkbench e.g. “DiversityWorkbench.Message.Connection.NoAccess”, PK |
| DisplayGroup |
nvarchar (50) |
If DiversityWorkbench entities should be displayed in a group, the name of the group |
| DisplayText |
nvarchar (50) |
Short abbreviated description of the object, displayed in the user interface |
| Abbreviation |
nvarchar (20) |
The abbreviation for the entity as shown e.g. in a user interface |
| Description |
nvarchar (MAX) |
Description of enumerated object, displayed in the user interface |
| Accessibility |
nvarchar (50) |
If the access of entity is resticted to e.g. read only or it can be edited without restrictions |
| Determination |
nvarchar (50) |
If a value is determined e.g. by the system or the user |
| Visibility |
nvarchar (50) |
If the entity is visible or hidden from e.g. a user interface |
| PresetValue |
nvarchar (500) |
If a value is preset, the value or SQL statement for the value, e.g. ‘determination’ for identifications when using a mobile device during an expedition |
| UsageNotes |
nvarchar (4000) |
Notes from table EntityUsage is present |
| DoesExist |
bit |
If an object does exist |
| DisplayTextOK |
bit |
If DisplayText is present |
| AbbreviationOK |
bit |
If Abbreviation is present |
| DescriptionOK |
bit |
If Description is present |
Depending on:
- Entity
- EntityContext_Enum
- EntityRepresentation
- EntityUsage
Function EventDescription
retrieval of the description including all superior events
DataType: nvarchar (4000)
| Parameter |
DataType |
Description |
| @CollectionEventID |
int |
Unique ID for the table CollectionEvent (= primary key) |
Depending on:
- CollectionEvent
- EventDescription
Function EventDescriptionSuperior
retrieval of the description of all superior events
DataType: nvarchar (4000)
| Parameter |
DataType |
Description |
| @CollectionEventID |
int |
Unique ID for the table CollectionEvent (= primary key) |
Depending on:
- CollectionEvent
- EventDescription
Function EventSeriesChildNodes
Returns a result set that lists all the items within a hierarchy starting at the topmost item related to the given item
| Parameter |
DataType |
Description |
| @ID |
int |
The SeriesID for which the children should be returned |
| Column |
DataType |
Description |
| SeriesID |
int |
Primary key. The ID for this series of collection events (= primary key) |
| SeriesParentID |
int |
The ID of the superior series of collection events |
| DateStart |
datetime |
Point in time when the series of collection events started |
| DateEnd |
datetime |
Point in time when the series of collection events ended |
| DateSupplement |
nvarchar (100) |
Verbal or additional collection date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. The end date, if the collection event series comprises a period. The time of the event, if necessary. |
| SeriesCode |
nvarchar (50) |
The user defined code for a series of collection events |
| Description |
nvarchar (MAX) |
The description of the series of collection events as it will be printed on e.g. the label |
| Notes |
nvarchar (MAX) |
Notes on this series of collection events |
| Geography |
geography (MAX) |
The geography of the series of collection events |
Depending on:
Function EventSeriesHierarchy
Returns a table that lists all the Series related to the given Series
| Parameter |
DataType |
Description |
| @SeriesID |
int |
Primary key. The ID for this series of collection events (= primary key) |
| Column |
DataType |
Description |
| SeriesID |
int |
Primary key. The ID for this series of collection events (= primary key) |
| SeriesParentID |
int |
The ID of the superior series of collection events |
| DateStart |
datetime |
Point in time when the series of collection events started |
| DateEnd |
datetime |
Point in time when the series of collection events ended |
| DateSupplement |
nvarchar (100) |
Verbal or additional collection date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. The end date, if the collection event series comprises a period. The time of the event, if necessary. |
| SeriesCode |
nvarchar (50) |
The user defined code for a series of collection events |
| Description |
nvarchar (MAX) |
The description of the series of collection events as it will be printed on e.g. the label |
| Notes |
nvarchar (MAX) |
Notes on this series of collection events |
| Geography |
geography (MAX) |
The geography of the series of collection events |
Depending on:
- CollectionEventSeries
- EventSeriesChildNodes
- EventSeriesTopID
Function EventSeriesSuperiorList
Returns a table that lists all the Series above the given Series.
| Parameter |
DataType |
Description |
| @SeriesID |
int |
Primary key. The ID for this series of collection events (= primary key) |
| Column |
DataType |
Description |
| SeriesID |
int |
Primary key. The ID for this series of collection events (= primary key) |
| SeriesParentID |
int |
The ID of the superior series of collection events |
| DateStart |
datetime |
Point in time when the series of collection events started |
| DateEnd |
datetime |
Point in time when the series of collection events ended |
| DateSupplement |
nvarchar (100) |
Verbal or additional collection date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. The end date, if the collection event series comprises a period. The time of the event, if necessary. |
| SeriesCode |
nvarchar (50) |
The user defined code for a series of collection events |
| Description |
nvarchar (500) |
The description of the series of collection events as it will be printed on e.g. the label |
| Notes |
nvarchar (500) |
Notes on this series of collection events |
| Geography |
geography (MAX) |
The geography of the series of collection events |
Depending on:
Function EventSeriesTopID
Returns the top ID within the hierarchy for a given ID from the table CollectionEventSeries
DataType: int
| Parameter |
DataType |
Description |
| @SeriesID |
int |
Primary key. The ID for this series of collection events (= primary key) |
Depending on:
Function EventSpecimenNumber
calculation of all specimen in the database that are assigned to this event, including the inferior events
DataType: int
| Parameter |
DataType |
Description |
| @EventID |
int |
The CollectionEventID for which the count should be calculated |
Depending on:
Function EventSuperiorList
Returns a result set that lists all the CollectionEventIDs superior to a given event
| Parameter |
DataType |
Description |
| @EventID |
int |
- |
| Column |
DataType |
Description |
| CollectionEventID |
int |
Refers to the ID of table CollectionEvent (= foreign key and part of primary key) |
| EventParentID |
int |
- |
| Event |
nchar (500) |
- |
| SpecimenCount |
int |
- |
Depending on:
Function FirstLines_4
Returns a table that lists all the specimen with the first entries of related tables
| Parameter |
DataType |
Description |
| @CollectionSpecimenIDs |
varchar (8000) |
Comma separated list of CollectionSpecimenIDs for which the data should be retrieved |
| Column |
DataType |
Description |
| CollectionSpecimenID |
int |
Refers to ID of CollectionEvent (= foreign key and part of primary key) |
| Accession_number |
nvarchar (50) |
Accession number of the specimen within the collection, e.g. “M-29834752” |
| Data_withholding_reason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
| Data_withholding_reason_for_collection_event |
nvarchar (255) |
- |
| Data_withholding_reason_for_collector |
nvarchar (255) |
If the data of the collector are withhold, the reason for withholding the data, otherwise null |
| Collectors_event_number |
nvarchar (50) |
Number assigned to a collection event by the collector (= ‘field number’) |
| Collection_day |
tinyint |
The day of the date of the event or when the collection event started |
| Collection_month |
tinyint |
The month of the date of the event or when the collection event started |
| Collection_year |
smallint |
The year of the date of the event or when the collection event started |
| Collection_date_supplement |
nvarchar (100) |
Verbal or additional collection date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. The end date, if the collection event comprises a period. The time of the event, if necessary. |
| Collection_time |
varchar (50) |
The time of the event or when the collection event started |
| Collection_time_span |
varchar (50) |
The time span e.g. in seconds of the collection event |
| Country |
nvarchar (50) |
The country where the collection event took place. Cached value derived from an geographic entry |
| Locality_description |
nvarchar (MAX) |
Locality description of the locality exactly as written on the original label (i.e. without corrections during data entry) |
| Habitat_description |
nvarchar (MAX) |
Geo-ecological description of the locality exactly as written on the original label (i.e. without corrections during data entry) |
| Collecting_method |
nvarchar (MAX) |
Description of the method used for collecting the samples, e.g. traps, moist chambers, drag net |
| Collection_event_notes |
nvarchar (MAX) |
Notes about the collection event |
| Named_area |
nvarchar (255) |
Named area as e.g. derived from DiversityGazetteer corresponding to LocalisationSystemID = 7 |
| NamedAreaLocation2 |
nvarchar (255) |
Link to a named area as e.g. derived from DiversityGazetteer corresponding to LocalisationSystemID = 7 |
| Remove_link_to_gazetteer |
int |
Empty column reserved to remove a link to a named area as e.g. derived from DiversityGazetteer corresponding to LocalisationSystemID = 7 |
| Distance_to_location |
varchar (50) |
Distance from the specified place to the real location of the collection site (m) |
| Direction_to_location |
varchar (50) |
Direction from the specified place to the real location of the collection site (Degrees rel. to north) |
| Longitude |
nvarchar (255) |
Longitude derived from WGS84 corresponding to LocalisationSystemID = 8 |
| Latitude |
nvarchar (255) |
Latitude derived from WGS84 corresponding to LocalisationSystemID = 8 |
| Coordinates_accuracy |
nvarchar (50) |
Accuracy of coordinates derived from WGS84 corresponding to LocalisationSystemID = 8 |
| Link_to_GoogleMaps |
int |
Empty column reserved for retrieval of WGS84 coordinates via e.g. GoogleMaps corresponding to LocalisationSystemID = 8 |
| _CoordinatesLocationNotes |
nvarchar (MAX) |
Notes for coordinates derived from WGS84 corresponding to LocalisationSystemID = 8 |
| Altitude_from |
nvarchar (255) |
Lower value of altitude range corresponding to LocalisationSystemID = 4 |
| Altitude_to |
nvarchar (255) |
Upper value of altitude range corresponding to LocalisationSystemID = 4 |
| Altitude_accuracy |
nvarchar (50) |
Accuracy of altitude range corresponding to LocalisationSystemID = 4 |
| Notes_for_Altitude |
nvarchar (MAX) |
Notes for altitude range corresponding to LocalisationSystemID = 4 |
| MTB |
nvarchar (255) |
TK25 corresponding to LocalisationSystemID = 3 |
| Quadrant |
nvarchar (255) |
Quadrant of TK25 corresponding to LocalisationSystemID = 3 |
| Notes_for_MTB |
nvarchar (MAX) |
Notes for TK25 corresponding to LocalisationSystemID = 3 |
| Sampling_plot |
nvarchar (255) |
Name of the sampling plot corresponding to LocalisationSystemID = 13 |
| Link_to_SamplingPlots |
nvarchar (255) |
Link to the sampling plot corresponding to LocalisationSystemID = 13 |
| Remove_link_to_SamplingPlots |
int |
Empty column reserverd to remove link to the sampling plot corresponding to LocalisationSystemID = 13 |
| Accuracy_of_sampling_plot |
nvarchar (50) |
Accuracy of the sampling plot coordinates corresponding to LocalisationSystemID = 13 |
| Latitude_of_sampling_plot |
real |
Latitude of the sampling plot coordinates corresponding to LocalisationSystemID = 13 |
| Longitude_of_sampling_plot |
real |
Longitude of the sampling plot coordinates corresponding to LocalisationSystemID = 13 |
| Geographic_region |
nvarchar (255) |
Geographic region corresponding to LocalisationSystemID = 10 |
| Lithostratigraphy |
nvarchar (255) |
Lithostratigraphy corresponding to LocalisationSystemID = 30 |
| Chronostratigraphy |
nvarchar (255) |
Chronostratigraphy corresponding to LocalisationSystemID = 20 |
| Biostratigraphy |
nvarchar (255) |
Biostratigraphy corresponding to LocalisationSystemID = 60 |
| Collectors_name |
nvarchar (255) |
Name of the Collector |
| Link_to_DiversityAgents |
varchar (255) |
Link for the first collector to DiversityAgents |
| Remove_link_for_collector |
int |
Empty column reserved for removal of ink for the first collector |
| Collectors_number |
nvarchar (50) |
Number assigned to a specimen or a batch of specimens by the collector during the collection event (= ‘field number’) |
| Notes_about_collector |
nvarchar (MAX) |
Notes about the first collector |
| Accession_day |
tinyint |
The day of the date when the specimen was acquired in the collection |
| Accession_month |
tinyint |
The month of the date when the specimen was acquired in the collection |
| Accession_year |
smallint |
The year of the date when the specimen was acquired in the collection |
| Accession_date_supplement |
nvarchar (255) |
Verbal or additional accession date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
| Depositors_name |
nvarchar (255) |
The name of the depositor(s) (person or organization responsible for deposition). Where entire collections are deposited, this should also contain the collection name (e.g. ‘Herbarium P. Döbbler’) |
| Depositors_link_to_DiversityAgents |
varchar (255) |
The link for the depositor e.g. to DiversityAgents |
| Remove_link_for_Depositor |
int |
Empty column reserved to remove the link for the depositor e.g. to DiversityAgents |
| Depositors_accession_number |
nvarchar (50) |
Accession number of the specimen within the previous or original collection, e.g. ‘D-23948’ |
| Exsiccata_abbreviation |
nvarchar (255) |
If specimen is an exsiccata: Standard abbreviation of the exsiccata (not necessarily a unique identifier; editors or publication places may change over time) |
| Link_to_DiversityExsiccatae |
varchar (255) |
Link to DiversityExsiccatae |
| Remove_link_to_exsiccatae |
int |
Empty column reserved to remove the link to DiversityExsiccatae |
| Exsiccata_number |
nvarchar (50) |
If specimen is an exsiccata: Number of current specimen within the exsiccata series |
| Original_notes |
nvarchar (MAX) |
Notes found on the label of the specimen by the original collector or from a later revision |
| Additional_notes |
nvarchar (MAX) |
Additional notes made by the editor of the specimen record, e.g. ‘doubtful identification/locality’ |
| Internal_notes |
nvarchar (MAX) |
Internal notes which should not be published e.g. on websites |
| Label_title |
nvarchar (255) |
The title of the label e.g. for printing labels. |
| Label_type |
nvarchar (50) |
Printed, typewritten, typewritten with handwriting added, entirely in handwriting, etc. |
| Label_transcription_state |
nvarchar (50) |
The state of the transcription of a label into the database: ‘Not started’, ‘incomplete’, ‘complete’ |
| Label_transcription_notes |
nvarchar (255) |
User defined notes on the transcription of the label into the database |
| Problems |
nvarchar (255) |
Description of a problem which occurred during data editing. Typically these entries should be deleted after help has been obtained. Do not enter scientific problems here; use AdditionalNotes for such permanent problems! |
| External_datasource |
int |
External datasource ID |
| External_identifier |
nvarchar (100) |
The identifier of the external specimen as defined in the external data source |
| Taxonomic_group |
nvarchar (50) |
Taxonomic group the organism, identified by this unit, belongs to. Groups listed in table CollTaxonomicGroup_Enum (= foreign key) |
| Relation_type |
nvarchar (50) |
Type of the relation between the specimen (= foreign key, see table CollRelationType_Enum) |
| Colonised_substrate_part |
nvarchar (255) |
If a substrate association exists: part of the substrate which is affected in the interaction (e.g. ’leaves’, if a fungus is growing on the leaves of an infected plant) |
| Life_stage |
nvarchar (255) |
Examples: ‘II, III’ for spore generations of rusts or ‘seed’, ‘seedling’ etc. for higher plants |
| Gender |
nvarchar (50) |
The sex of the organism, e.g. ‘female’ |
| Number_of_units |
smallint |
The number of units of this organism, e.g. 400 beetles in a bottle |
| Circumstances |
nvarchar (50) |
Circumstances of the occurence of the organism |
| Order_of_taxon |
nvarchar (255) |
Order of the first taxon |
| Family_of_taxon |
nvarchar (255) |
Family of the first taxon |
| Identifier_of_organism |
nvarchar (50) |
Identifier of the first organism |
| Description_of_organism |
nvarchar (50) |
Description of the first organism |
| Only_observed |
bit |
True, if the organism was only observed rather than collected. It is therefore not present on the preserved specimen. Example: Tree under which the collected mycorrhizal fungus grew. |
| Notes_for_organism |
nvarchar (MAX) |
Notes for the first organism |
| Taxonomic_name |
nvarchar (255) |
Valid name of the species (including the taxonomic author where available). Example: ‘Rosa canina L.’ |
| Link_to_DiversityTaxonNames |
varchar (255) |
Link for first identification of first organism to DiversityTaxonNames |
| Remove_link_for_identification |
int |
Empty column to remove link for first identification of first organism to DiversityTaxonNames |
| Vernacular_term |
nvarchar (255) |
Name or term other than a taxonomic (= scientific) name, e.g. ‘pine’, ’limestone’, ‘conifer’, ‘hardwood’ |
| Identification_day |
tinyint |
The day of the identification |
| Identification_month |
tinyint |
The month of the identification |
| Identification_year |
smallint |
The year of the identification. The year may be empty if only the day or month are known. |
| Identification_category |
nvarchar (50) |
Category of the identification e.g. ‘determination’, ‘confirmation’, ‘absence’ (= foreign key, see table CollIdentificationCategory_Enum) |
| Identification_qualifier |
nvarchar (50) |
Qualification of the identification e.g. “cf.”," aff.", “sp. nov.” (= foreign key, see table CollIdentificationQualifier_Enum) |
| Type_status |
nvarchar (50) |
If identification unit is type of a taxonomic name: holotype, syntype, etc. (= foreign key, see table CollTypeStatus_Enum) |
| Type_notes |
nvarchar (MAX) |
Notes on the typification of this specimen |
| Notes_for_identification |
nvarchar (MAX) |
Notes for first identification of first organism to DiversityTaxonNames |
| Determiner |
nvarchar (255) |
Agent responsible for first identification of first organism |
| Link_to_DiversityAgents_for_determiner |
varchar (255) |
Agent for responsible for first identification of first organism to e.g. DiversityAgents |
| Remove_link_for_determiner |
int |
Empty column reservet to remove link for agent for responsible for first identification of first organism to e.g. DiversityAgents |
| Analysis |
nvarchar (50) |
First analysis of first organism |
| AnalysisID |
int |
ID of the analysis (primary key) |
| Analysis_number |
nvarchar (50) |
Number of the analysis |
| Analysis_result |
nvarchar (MAX) |
The result of the analysis |
| Taxonomic_group_of_second_organism |
nvarchar (50) |
Taxonomic group of second organism |
| Life_stage_of_second_organism |
nvarchar (255) |
Life stage of second organism |
| Gender_of_second_organism |
nvarchar (50) |
Gender of second organism |
| Number_of_units_of_second_organism |
smallint |
Number of units of second organism |
| Circumstances_of_second_organism |
nvarchar (50) |
Circumstances of second organism |
| Identifier_of_second_organism |
nvarchar (50) |
Identifier for second organism |
| Description_of_second_organism |
nvarchar (50) |
UnitDescription of second organism |
| Only_observed_of_second_organism |
bit |
OnlyObserved of second organism |
| Notes_for_second_organism |
nvarchar (MAX) |
Notes for second organism |
| Taxonomic_name_of_second_organism |
nvarchar (255) |
Taxonomic name of second organism |
| Link_to_DiversityTaxonNames_of_second_organism |
varchar (255) |
Link to DiversityTaxonNames of second organism |
| Remove_link_for_second_organism |
int |
Remove link to DiversityTaxonNames for second organism |
| Vernacular_term_of_second_organism |
nvarchar (255) |
Vernacular term of second organism |
| Identification_day_of_second_organism |
tinyint |
Identification day of second organism |
| Identification_month_of_second_organism |
tinyint |
Identification month of second organism |
| Identification_year_of_second_organism |
smallint |
Identification year of second organism |
| Identification_category_of_second_organism |
nvarchar (50) |
- |
| Identification_qualifier_of_second_organism |
nvarchar (50) |
- |
| Type_status_of_second_organism |
nvarchar (50) |
- |
| Type_notes_of_second_organism |
nvarchar (MAX) |
- |
| Notes_for_identification_of_second_organism |
nvarchar (MAX) |
- |
| Determiner_of_second_organism |
nvarchar (255) |
- |
| Link_to_DiversityAgents_for_determiner_of_second_organism |
varchar (255) |
- |
| Remove_link_for_determiner_of_second_organism |
int |
- |
| Collection |
int |
- |
| Material_category |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. (= foreign key, see table CollMaterialCategory_Enum) |
| Storage_location |
nvarchar (255) |
A code identifying the place where the specimen is stored within the collection. Frequently the accepted scientific name is used as storage location code. |
| Stock |
float |
Number of stock units, if the specimen is stored in separated units e.g. several boxes or vessels (max. 255) |
| Part_accession_number |
nvarchar (50) |
- |
| Storage_container |
nvarchar (500) |
The container in which the part is stored |
| Preparation_method |
nvarchar (MAX) |
The method used for the preparation of the part of the specimen, e.g. the inoculation method for cultures |
| Preparation_date |
datetime |
Point in time when the part was preparated e.g when it was separated from the source object |
| Notes_for_part |
nvarchar (MAX) |
- |
| Related_specimen_URL |
varchar (255) |
- |
| Related_specimen_display_text |
varchar (255) |
The name of a related specimen as shown e.g. in a user interface |
| Link_to_DiversityCollection_for_relation |
varchar (255) |
- |
| Type_of_relation |
nvarchar (50) |
- |
| Related_specimen_description |
nvarchar (MAX) |
Description of the related specimen |
| Related_specimen_notes |
nvarchar (MAX) |
- |
| Relation_is_internal |
bit |
- |
| _TransactionID |
int |
Unique ID for the table Transaction (= foreign key and part of primary key) |
| _Transaction |
nvarchar (200) |
- |
| _CollectionEventID |
int |
Unique ID for the table CollectionEvent (= primary key) |
| _IdentificationUnitID |
int |
If relation refers to a certain organism within a specimen, the ID of an IdentificationUnit (= foreign key) |
| _IdentificationSequence |
smallint |
The sequence of the identifications. The last identification (having the highest sequence) is regarded as valid |
| _SecondUnitID |
int |
- |
| _SecondSequence |
smallint |
- |
| _SpecimenPartID |
int |
Unique ID of the part of the collection specimen (= part of primary key). |
| _CoordinatesAverageLatitudeCache |
real |
- |
| _CoordinatesAverageLongitudeCache |
real |
- |
| _GeographicRegionPropertyURI |
varchar (255) |
- |
| _LithostratigraphyPropertyURI |
varchar (255) |
- |
| _ChronostratigraphyPropertyURI |
varchar (255) |
- |
| _BiostratigraphyPropertyURI |
varchar (255) |
- |
| _NamedAverageLatitudeCache |
real |
- |
| _NamedAverageLongitudeCache |
real |
- |
| _LithostratigraphyPropertyHierarchyCache |
nvarchar (MAX) |
- |
| _ChronostratigraphyPropertyHierarchyCache |
nvarchar (MAX) |
- |
| _BiostratigraphyPropertyHierarchyCache |
nvarchar (MAX) |
- |
| _SecondUnitFamilyCache |
nvarchar (255) |
- |
| _SecondUnitOrderCache |
nvarchar (255) |
- |
| _AverageAltitudeCache |
real |
Calculated altitude as parsed from the location fields |
Depending on:
- Analysis
- CollectionAgent
- CollectionEvent
- CollectionEventLocalisation
- CollectionEventProperty
- CollectionSpecimen
- CollectionSpecimenID_UserAvailable
- CollectionSpecimenPart
- CollectionSpecimenRelation
- CollectionSpecimenTransaction
- Identification
- IdentificationUnit
- IdentificationUnitAnalysis
- Transaction
Function FirstLinesEvent_2
Returns a table that lists all the collection event with the first entries of related tables
| Parameter |
DataType |
Description |
| @CollectionSpecimenIDs |
varchar (8000) |
Comma separated list of CollectionSpecimenIDs for which the data should be retrieved |
| Column |
DataType |
Description |
| CollectionEventID |
int |
Unique ID for the table CollectionEvent (= primary key) |
| Data_withholding_reason_for_collection_event |
nvarchar (255) |
If the data of the collection event are withhold, the reason for withholding the data, otherwise null |
| Collectors_event_number |
nvarchar (50) |
Number assigned to a collection event by the collector (= ‘field number’) |
| Collection_day |
tinyint |
The day of the date of the event or when the collection event started |
| Collection_month |
tinyint |
The month of the date of the event or when the collection event started |
| Collection_year |
smallint |
The year of the date of the event or when the collection event started |
| Collection_date_supplement |
nvarchar (100) |
Verbal or additional collection date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. The end date, if the collection event comprises a period. The time of the event, if necessary. |
| Collection_time |
varchar (50) |
The time of the event or when the collection event started |
| Collection_time_span |
varchar (50) |
The time span e.g. in seconds of the collection event |
| Country |
nvarchar (50) |
The country where the collection event took place. Cached value derived from an geographic entry |
| Locality_description |
nvarchar (MAX) |
Locality description of the locality exactly as written on the original label (i.e. without corrections during data entry) |
| Habitat_description |
nvarchar (MAX) |
Geo-ecological description of the locality exactly as written on the original label (i.e. without corrections during data entry) |
| Collecting_method |
nvarchar (MAX) |
Description of the method used for collecting the samples, e.g. traps, moist chambers, drag net |
| Collection_event_notes |
nvarchar (MAX) |
Notes about the collection event |
| Named_area |
nvarchar (255) |
Named area as e.g. derived from DiversityGazetteer corresponding to LocalisationSystemID = 7 |
| NamedAreaLocation2 |
nvarchar (255) |
Link to a named area as e.g. derived from DiversityGazetteer corresponding to LocalisationSystemID = 7 |
| Remove_link_to_gazetteer |
int |
Empty column reserved to remove a link to a named area as e.g. derived from DiversityGazetteer corresponding to LocalisationSystemID = 7 |
| Distance_to_location |
varchar (50) |
Distance from the specified place to the real location of the collection site (m) |
| Direction_to_location |
varchar (50) |
Direction from the specified place to the real location of the collection site (Degrees rel. to north) |
| Longitude |
nvarchar (255) |
Longitude derived from WGS84 corresponding to LocalisationSystemID = 8 |
| Latitude |
nvarchar (255) |
Latitude derived from WGS84 corresponding to LocalisationSystemID = 8 |
| Coordinates_accuracy |
nvarchar (50) |
Accuracy of coordinates derived from WGS84 corresponding to LocalisationSystemID = 8 |
| Link_to_GoogleMaps |
int |
Empty column reserved for retrieval of WGS84 coordinates via e.g. GoogleMaps corresponding to LocalisationSystemID = 8 |
| Altitude_from |
nvarchar (255) |
Lower value of altitude range corresponding to LocalisationSystemID = 4 |
| Altitude_to |
nvarchar (255) |
Upper value of altitude range corresponding to LocalisationSystemID = 4 |
| Altitude_accuracy |
nvarchar (50) |
Accuracy of altitude range corresponding to LocalisationSystemID = 4 |
| MTB |
nvarchar (255) |
TK25 corresponding to LocalisationSystemID = 3 |
| Quadrant |
nvarchar (255) |
Quadrant of TK25 corresponding to LocalisationSystemID = 3 |
| Notes_for_MTB |
nvarchar (MAX) |
Notes for TK25 corresponding to LocalisationSystemID = 3 |
| MTB_accuracy |
nvarchar (50) |
- |
| Sampling_plot |
nvarchar (255) |
Name of the sampling plot corresponding to LocalisationSystemID = 13 |
| Link_to_SamplingPlots |
nvarchar (255) |
Link to the sampling plot corresponding to LocalisationSystemID = 13 |
| Remove_link_to_SamplingPlots |
int |
Empty column reserverd to remove link to the sampling plot corresponding to LocalisationSystemID = 13 |
| Accuracy_of_sampling_plot |
nvarchar (50) |
Accuracy of the sampling plot coordinates corresponding to LocalisationSystemID = 13 |
| Latitude_of_sampling_plot |
real |
Latitude of the sampling plot coordinates corresponding to LocalisationSystemID = 13 |
| Longitude_of_sampling_plot |
real |
Longitude of the sampling plot coordinates corresponding to LocalisationSystemID = 13 |
| Geographic_region |
nvarchar (255) |
Geographic region corresponding to LocalisationSystemID = 10 |
| Lithostratigraphy |
nvarchar (255) |
Lithostratigraphy corresponding to LocalisationSystemID = 30 |
| Chronostratigraphy |
nvarchar (255) |
Chronostratigraphy corresponding to LocalisationSystemID = 20 |
| _CollectionEventID |
int |
Unique ID for the table CollectionEvent (= primary key) |
| _CoordinatesAverageLatitudeCache |
real |
Average Latitude of WGS84 Coordinates |
| _CoordinatesAverageLongitudeCache |
real |
Average Longitude of WGS84 Coordinates |
| _CoordinatesLocationNotes |
nvarchar (MAX) |
Notes for WGS84 Coordinates |
| _GeographicRegionPropertyURI |
varchar (255) |
URI of Geographic Region |
| _LithostratigraphyPropertyURI |
varchar (255) |
URI for Lithostratigraphy |
| _ChronostratigraphyPropertyURI |
varchar (255) |
URI for Chronostratigraphy |
| _NamedAverageLatitudeCache |
real |
Average Latitude for Named Area |
| _NamedAverageLongitudeCache |
real |
Average Longitude for Named Area |
| _LithostratigraphyPropertyHierarchyCache |
nvarchar (MAX) |
Hierarchy for Lithostratigraphy |
| _ChronostratigraphyPropertyHierarchyCache |
nvarchar (MAX) |
Hierarchy for Chronostratigraphy |
| _AverageAltitudeCache |
real |
Average Altitude |
Depending on:
- CollectionEvent
- CollectionEventLocalisation
- CollectionEventProperty
- CollectionSpecimen
- CollectionSpecimenID_UserAvailable
Function FirstLinesPart_2
Returns a table that lists all the collection specimen part with the first entries of related tables
| Parameter |
DataType |
Description |
| @CollectionSpecimenIDs |
varchar (8000) |
Comma separated list of CollectionSpecimenIDs for which the data should be retrieved |
| @AnalysisIDs |
varchar (8000) |
Comma separated list of AnalysisIDs for which the data should be retrieved |
| @AnalysisStartDate |
date |
The start date of the range for analysis |
| @AnalysisEndDate |
date |
The end date of the range for analysis |
| @ProcessingID |
int |
ID of the processing method. Refers to ProcessingID in table Processing (foreign key) |
| @ProcessingStartDate |
datetime |
The start date of the range for processing |
| @ProcessingEndDate |
datetime |
The end date of the range for processing |
| Column |
DataType |
Description |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen (= part of primary key). |
| CollectionSpecimenID |
int |
Refers to ID of CollectionEvent (= foreign key and part of primary key) |
| Accession_number |
nvarchar (50) |
Accession number of the specimen within the collection, e.g. “M-29834752” |
| Data_withholding_reason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
| Data_withholding_reason_for_collection_event |
nvarchar (255) |
If the data of the collection event are withhold, the reason for withholding the data, otherwise null |
| Data_withholding_reason_for_collector |
nvarchar (255) |
If the data of the collector are withhold, the reason for withholding the data, otherwise null |
| Collectors_event_number |
nvarchar (50) |
Number assigned to a collection event by the collector (= ‘field number’) |
| Collection_day |
tinyint |
The day of the date of the event or when the collection event started |
| Collection_month |
tinyint |
The month of the date of the event or when the collection event started |
| Collection_year |
smallint |
The year of the date of the event or when the collection event started |
| Collection_date_supplement |
nvarchar (100) |
Verbal or additional collection date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. The end date, if the collection event comprises a period. The time of the event, if necessary. |
| Collection_time |
varchar (50) |
The time of the event or when the collection event started |
| Collection_time_span |
varchar (50) |
The time span e.g. in seconds of the collection event |
| Country |
nvarchar (50) |
The country where the collection event took place. Cached value derived from an geographic entry |
| Locality_description |
nvarchar (255) |
Locality description of the locality exactly as written on the original label (i.e. without corrections during data entry) |
| Habitat_description |
nvarchar (255) |
Geo-ecological description of the locality exactly as written on the original label (i.e. without corrections during data entry) |
| Collecting_method |
nvarchar (255) |
Description of the method used for collecting the samples, e.g. traps, moist chambers, drag net |
| Collection_event_notes |
nvarchar (255) |
Notes about the collection event |
| Named_area |
nvarchar (255) |
Named area as e.g. derived from DiversityGazetteer corresponding to LocalisationSystemID = 7 |
| NamedAreaLocation2 |
nvarchar (255) |
Link to a named area as e.g. derived from DiversityGazetteer corresponding to LocalisationSystemID = 7 |
| Remove_link_to_gazetteer |
int |
Empty column reserved to remove a link to a named area as e.g. derived from DiversityGazetteer corresponding to LocalisationSystemID = 7 |
| Distance_to_location |
varchar (50) |
Distance from the specified place to the real location of the collection site (m) |
| Direction_to_location |
varchar (50) |
Direction from the specified place to the real location of the collection site (Degrees rel. to north) |
| Longitude |
nvarchar (255) |
Longitude derived from WGS84 corresponding to LocalisationSystemID = 8 |
| Latitude |
nvarchar (255) |
Latitude derived from WGS84 corresponding to LocalisationSystemID = 8 |
| Coordinates_accuracy |
nvarchar (50) |
Accuracy of coordinates derived from WGS84 corresponding to LocalisationSystemID = 8 |
| Link_to_GoogleMaps |
int |
Empty column reserved for retrieval of WGS84 coordinates via e.g. GoogleMaps corresponding to LocalisationSystemID = 8 |
| Altitude_from |
nvarchar (255) |
Lower value of altitude range corresponding to LocalisationSystemID = 4 |
| Altitude_to |
nvarchar (255) |
Upper value of altitude range corresponding to LocalisationSystemID = 4 |
| Altitude_accuracy |
nvarchar (50) |
Accuracy of altitude range corresponding to LocalisationSystemID = 4 |
| Notes_for_Altitude |
nvarchar (255) |
Notes for altitude range corresponding to LocalisationSystemID = 4 |
| MTB |
nvarchar (255) |
TK25 corresponding to LocalisationSystemID = 3 |
| Quadrant |
nvarchar (255) |
Quadrant of TK25 corresponding to LocalisationSystemID = 3 |
| Notes_for_MTB |
nvarchar (255) |
Notes for TK25 corresponding to LocalisationSystemID = 3 |
| Sampling_plot |
nvarchar (255) |
Name of the sampling plot corresponding to LocalisationSystemID = 13 |
| Link_to_SamplingPlots |
nvarchar (255) |
Link to the sampling plot corresponding to LocalisationSystemID = 13 |
| Remove_link_to_SamplingPlots |
int |
Empty column reserverd to remove link to the sampling plot corresponding to LocalisationSystemID = 13 |
| Accuracy_of_sampling_plot |
nvarchar (50) |
Accuracy of the sampling plot coordinates corresponding to LocalisationSystemID = 13 |
| Latitude_of_sampling_plot |
real |
Latitude of the sampling plot coordinates corresponding to LocalisationSystemID = 13 |
| Longitude_of_sampling_plot |
real |
Longitude of the sampling plot coordinates corresponding to LocalisationSystemID = 13 |
| Geographic_region |
nvarchar (255) |
Geographic region corresponding to LocalisationSystemID = 10 |
| Lithostratigraphy |
nvarchar (255) |
Lithostratigraphy corresponding to LocalisationSystemID = 30 |
| Chronostratigraphy |
nvarchar (255) |
Chronostratigraphy corresponding to LocalisationSystemID = 20 |
| Collectors_name |
nvarchar (255) |
Name of the Collector |
| Link_to_DiversityAgents |
varchar (255) |
Link for the first collector to DiversityAgents |
| Remove_link_for_collector |
int |
- |
| Collectors_number |
nvarchar (50) |
Number assigned to a specimen or a batch of specimens by the collector during the collection event (= ‘field number’) |
| Notes_about_collector |
nvarchar (MAX) |
Notes about the first collector |
| Accession_day |
tinyint |
The day of the date when the specimen was acquired in the collection |
| Accession_month |
tinyint |
The month of the date when the specimen was acquired in the collection |
| Accession_year |
smallint |
The year of the date when the specimen was acquired in the collection |
| Accession_date_supplement |
nvarchar (255) |
Verbal or additional accession date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
| Depositors_name |
nvarchar (255) |
The name of the depositor(s) (person or organization responsible for deposition). Where entire collections are deposited, this should also contain the collection name (e.g. ‘Herbarium P. Döbbler’) |
| Depositors_link_to_DiversityAgents |
varchar (255) |
The link for the depositor e.g. to DiversityAgents |
| Remove_link_for_Depositor |
int |
Empty column reserved to remove the link for the depositor e.g. to DiversityAgents |
| Depositors_accession_number |
nvarchar (50) |
Accession number of the specimen within the previous or original collection, e.g. ‘D-23948’ |
| Exsiccata_abbreviation |
nvarchar (255) |
If specimen is an exsiccata: Standard abbreviation of the exsiccata (not necessarily a unique identifier; editors or publication places may change over time) |
| Link_to_DiversityExsiccatae |
varchar (255) |
Link to DiversityExsiccatae |
| Remove_link_to_exsiccatae |
int |
Empty column reserved to remove the link to DiversityExsiccatae |
| Exsiccata_number |
nvarchar (50) |
If specimen is an exsiccata: Number of current specimen within the exsiccata series |
| Original_notes |
nvarchar (MAX) |
Notes found on the label of the specimen by the original collector or from a later revision |
| Additional_notes |
nvarchar (MAX) |
Additional notes made by the editor of the specimen record, e.g. ‘doubtful identification/locality’ |
| Internal_notes |
nvarchar (MAX) |
Internal notes which should not be published e.g. on websites |
| Label_title |
nvarchar (255) |
The title of the label e.g. for printing labels. |
| Label_type |
nvarchar (50) |
Printed, typewritten, typewritten with handwriting added, entirely in handwriting, etc. |
| Label_transcription_state |
nvarchar (50) |
The state of the transcription of a label into the database: ‘Not started’, ‘incomplete’, ‘complete’ |
| Label_transcription_notes |
nvarchar (255) |
User defined notes on the transcription of the label into the database |
| Problems |
nvarchar (255) |
Description of a problem which occurred during data editing. Typically these entries should be deleted after help has been obtained. Do not enter scientific problems here; use AdditionalNotes for such permanent problems! |
| Taxonomic_group |
nvarchar (50) |
Taxonomic group the organism, identified by this unit, belongs to. Groups listed in table CollTaxonomicGroup_Enum (= foreign key) |
| Relation_type |
nvarchar (50) |
The relation of a unit to its substrate, e.g. parasitism, symbiosis etc. as stored in CollRelationType_Enum (= foreign key) |
| Colonised_substrate_part |
nvarchar (255) |
If a substrate association exists: part of the substrate which is affected in the interaction (e.g. ’leaves’, if a fungus is growing on the leaves of an infected plant) |
| Related_organism |
nvarchar (200) |
Last identification of the related organism |
| Life_stage |
nvarchar (255) |
Examples: ‘II, III’ for spore generations of rusts or ‘seed’, ‘seedling’ etc. for higher plants |
| Gender |
nvarchar (50) |
The sex of the organism, e.g. ‘female’ |
| Number_of_units |
smallint |
The number of units of this organism, e.g. 400 beetles in a bottle |
| Circumstances |
nvarchar (50) |
Circumstances of the occurence of the organism |
| Order_of_taxon |
nvarchar (255) |
Order of the first taxon |
| Family_of_taxon |
nvarchar (255) |
Family of the first taxon |
| Identifier_of_organism |
nvarchar (50) |
Identifier of the first organism |
| Description_of_organism |
nvarchar (50) |
Description of the first organism |
| Only_observed |
bit |
True, if the organism was only observed rather than collected. It is therefore not present on the preserved specimen. Example: Tree under which the collected mycorrhizal fungus grew. |
| Notes_for_organism |
nvarchar (MAX) |
Notes for the first organism |
| Taxonomic_name |
nvarchar (255) |
Valid name of the species (including the taxonomic author where available). Example: ‘Rosa canina L.’ |
| Link_to_DiversityTaxonNames |
varchar (255) |
Link for first identification of first organism to DiversityTaxonNames |
| Remove_link_for_identification |
int |
Empty column to remove link for first identification of first organism to DiversityTaxonNames |
| Vernacular_term |
nvarchar (255) |
Name or term other than a taxonomic (= scientific) name, e.g. ‘pine’, ’limestone’, ‘conifer’, ‘hardwood’ |
| Identification_day |
tinyint |
The day of the identification |
| Identification_month |
tinyint |
The month of the identification |
| Identification_year |
smallint |
The year of the identification. The year may be empty if only the day or month are known. |
| Identification_category |
nvarchar (50) |
Category of the identification e.g. ‘determination’, ‘confirmation’, ‘absence’ (= foreign key, see table CollIdentificationCategory_Enum) |
| Identification_qualifier |
nvarchar (50) |
Qualification of the identification e.g. “cf.”," aff.", “sp. nov.” (= foreign key, see table CollIdentificationQualifier_Enum) |
| Type_status |
nvarchar (50) |
If identification unit is type of a taxonomic name: holotype, syntype, etc. (= foreign key, see table CollTypeStatus_Enum) |
| Type_notes |
nvarchar (MAX) |
Notes on the typification of this specimen |
| Notes_for_identification |
nvarchar (MAX) |
Notes for first identification of first organism to DiversityTaxonNames |
| Determiner |
nvarchar (255) |
Agent responsible for first identification of first organism |
| Link_to_DiversityAgents_for_determiner |
varchar (255) |
Agent for responsible for first identification of first organism to e.g. DiversityAgents |
| Remove_link_for_determiner |
int |
Empty column reservet to remove link for agent for responsible for first identification of first organism to e.g. DiversityAgents |
| Analysis_0 |
nvarchar (50) |
Name of analysis 0 according to list of AnalysisIDs |
| AnalysisID_0 |
int |
ID of analysis 0 according to list of AnalysisIDs |
| Analysis_number_0 |
nvarchar (50) |
Number of analysis 0 according to list of AnalysisIDs |
| Analysis_result_0 |
nvarchar (MAX) |
Result of analysis 0 according to list of AnalysisIDs |
| Analysis_1 |
nvarchar (50) |
Name of analysis_1 according to list of AnalysisIDs |
| AnalysisID_1 |
int |
ID of analysis_1 according to list of AnalysisIDs |
| Analysis_number_1 |
nvarchar (50) |
Number of analysis_1 according to list of AnalysisIDs |
| Analysis_result_1 |
nvarchar (MAX) |
Result of analysis_1 according to list of AnalysisIDs |
| Analysis_2 |
nvarchar (50) |
Name of analysis_2 according to list of AnalysisIDs |
| AnalysisID_2 |
int |
ID of analysis_2 according to list of AnalysisIDs |
| Analysis_number_2 |
nvarchar (50) |
Number of analysis_2 according to list of AnalysisIDs |
| Analysis_result_2 |
nvarchar (MAX) |
Result of analysis_2 according to list of AnalysisIDs |
| Analysis_3 |
nvarchar (50) |
Name of analysis_3 according to list of AnalysisIDs |
| AnalysisID_3 |
int |
ID of analysis_3 according to list of AnalysisIDs |
| Analysis_number_3 |
nvarchar (50) |
Number of analysis_3 according to list of AnalysisIDs |
| Analysis_result_3 |
nvarchar (MAX) |
Result of analysis_3 according to list of AnalysisIDs |
| Analysis_4 |
nvarchar (50) |
Name of analysis_4 according to list of AnalysisIDs |
| AnalysisID_4 |
int |
ID of analysis_4 according to list of AnalysisIDs |
| Analysis_number_4 |
nvarchar (50) |
Number of analysis_4 according to list of AnalysisIDs |
| Analysis_result_4 |
nvarchar (MAX) |
Result of analysis_4 according to list of AnalysisIDs |
| Analysis_5 |
nvarchar (50) |
Name of analysis_5 according to list of AnalysisIDs |
| AnalysisID_5 |
int |
ID of analysis_5 according to list of AnalysisIDs |
| Analysis_number_5 |
nvarchar (50) |
Number of analysis_5 according to list of AnalysisIDs |
| Analysis_result_5 |
nvarchar (MAX) |
Result of analysis_5 according to list of AnalysisIDs |
| Analysis_6 |
nvarchar (50) |
Name of analysis_6 according to list of AnalysisIDs |
| AnalysisID_6 |
int |
ID of analysis_6 according to list of AnalysisIDs |
| Analysis_number_6 |
nvarchar (50) |
Number of analysis_6 according to list of AnalysisIDs |
| Analysis_result_6 |
nvarchar (MAX) |
Result of analysis_6 according to list of AnalysisIDs |
| Analysis_7 |
nvarchar (50) |
Name of analysis_7 according to list of AnalysisIDs |
| AnalysisID_7 |
int |
ID of analysis_7 according to list of AnalysisIDs |
| Analysis_number_7 |
nvarchar (50) |
Number of analysis_7 according to list of AnalysisIDs |
| Analysis_result_7 |
nvarchar (MAX) |
Result of analysis_7 according to list of AnalysisIDs |
| Analysis_8 |
nvarchar (50) |
Name of analysis_8 according to list of AnalysisIDs |
| AnalysisID_8 |
int |
ID of analysis_8 according to list of AnalysisIDs |
| Analysis_number_8 |
nvarchar (50) |
Number of analysis_8 according to list of AnalysisIDs |
| Analysis_result_8 |
nvarchar (MAX) |
Result of analysis_8 according to list of AnalysisIDs |
| Analysis_9 |
nvarchar (50) |
Name of analysis_9 according to list of AnalysisIDs |
| AnalysisID_9 |
int |
ID of analysis_9 according to list of AnalysisIDs |
| Analysis_number_9 |
nvarchar (50) |
Number of analysis_9 according to list of AnalysisIDs |
| Analysis_result_9 |
nvarchar (MAX) |
Result of analysis_9 according to list of AnalysisIDs |
| Preparation_method |
nvarchar (MAX) |
The method used for the preparation of the part of the specimen, e.g. the inoculation method for cultures |
| Preparation_date |
datetime |
Point in time when the part was preparated e.g when it was separated from the source object |
| Part_accession_number |
nvarchar (50) |
Accession number of part |
| Part_sublabel |
nvarchar (50) |
The label for a part of a specimen, e.g. “cone”, or a number attached to a duplicate of a specimen |
| Collection |
int |
ID of the Collection |
| Material_category |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. (= foreign key, see table CollMaterialCategory_Enum) |
| Storage_location |
nvarchar (255) |
A code identifying the place where the specimen is stored within the collection. Frequently the accepted scientific name is used as storage location code. |
| Storage_container |
nvarchar (500) |
The container in which the part is stored |
| Stock |
float |
Number of stock units, if the specimen is stored in separated units e.g. several boxes or vessels (max. 255) |
| Stock_unit |
nvarchar (50) |
If empty, the stock is given as a count, else it contains the unit in which stock is expressed, e.g. µl, ml, kg etc. |
| Notes_for_part |
nvarchar (MAX) |
Notes for specimen part |
| Description_of_unit_in_part |
nvarchar (500) |
Description of identification unit in specimen part |
| Processing_date_1 |
datetime |
Date of Processing_1 according to list of ProcessingIDs |
| ProcessingID_1 |
int |
ID of Processing_1 according to list of ProcessingIDs |
| Processing_Protocoll_1 |
nvarchar (100) |
Protocoll of Processing_1 according to list of ProcessingIDs |
| Processing_duration_1 |
varchar (50) |
Duration of Processing_1 according to list of ProcessingIDs |
| Processing_notes_1 |
nvarchar (MAX) |
Notes of Processing_1 according to list of ProcessingIDs |
| Processing_date_2 |
datetime |
Date of Processing_2 according to list of ProcessingIDs |
| ProcessingID_2 |
int |
ID of Processing_2 according to list of ProcessingIDs |
| Processing_Protocoll_2 |
nvarchar (100) |
Protocoll of Processing_2 according to list of ProcessingIDs |
| Processing_duration_2 |
varchar (50) |
Duration of Processing_2 according to list of ProcessingIDs |
| Processing_notes_2 |
nvarchar (MAX) |
Notes of Processing_2 according to list of ProcessingIDs |
| Processing_date_3 |
datetime |
Date of Processing_3 according to list of ProcessingIDs |
| ProcessingID_3 |
int |
ID of Processing_3 according to list of ProcessingIDs |
| Processing_Protocoll_3 |
nvarchar (100) |
Protocoll of Processing_3 according to list of ProcessingIDs |
| Processing_duration_3 |
varchar (50) |
Duration of Processing_3 according to list of ProcessingIDs |
| Processing_notes_3 |
nvarchar (MAX) |
Notes of Processing_3 according to list of ProcessingIDs |
| Processing_date_4 |
datetime |
Date of Processing_4 according to list of ProcessingIDs |
| ProcessingID_4 |
int |
ID of Processing_4 according to list of ProcessingIDs |
| Processing_Protocoll_4 |
nvarchar (100) |
Protocoll of Processing_4 according to list of ProcessingIDs |
| Processing_duration_4 |
varchar (50) |
Duration of Processing_4 according to list of ProcessingIDs |
| Processing_notes_4 |
nvarchar (MAX) |
Notes of Processing_4 according to list of ProcessingIDs |
| Processing_date_5 |
datetime |
Date of Processing_5 according to list of ProcessingIDs |
| ProcessingID_5 |
int |
ID of Processing_5 according to list of ProcessingIDs |
| Processing_Protocoll_5 |
nvarchar (100) |
Protocoll of Processing_5 according to list of ProcessingIDs |
| Processing_duration_5 |
varchar (50) |
Duration of Processing_5 according to list of ProcessingIDs |
| Processing_notes_5 |
nvarchar (MAX) |
Notes of Processing_5 according to list of ProcessingIDs |
| _TransactionID |
int |
Unique ID for the table Transaction (= foreign key and part of primary key) |
| _Transaction |
nvarchar (200) |
Title of first transaction |
| On_loan |
int |
If part is on loan according to first transaction |
| _CollectionEventID |
int |
Unique ID for the table CollectionEvent (= primary key) |
| _IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
| _IdentificationSequence |
smallint |
The sequence of the identifications. The last identification (having the highest sequence) is regarded as valid |
| _SpecimenPartID |
int |
Unique ID of the part of the collection specimen (= part of primary key). |
| _CoordinatesAverageLatitudeCache |
real |
Average Latitude of WGS84 Coordinates |
| _CoordinatesAverageLongitudeCache |
real |
Average Longitude of WGS84 Coordinates |
| _CoordinatesLocationNotes |
nvarchar (255) |
Notes for WGS84 Coordinates |
| _GeographicRegionPropertyURI |
varchar (255) |
URI of Geographic Region |
| _LithostratigraphyPropertyURI |
varchar (255) |
URI for Lithostratigraphy |
| _ChronostratigraphyPropertyURI |
varchar (255) |
URI for Chronostratigraphy |
| _NamedAverageLatitudeCache |
real |
Average Latitude for Named Area |
| _NamedAverageLongitudeCache |
real |
Average Longitude for Named Area |
| _LithostratigraphyPropertyHierarchyCache |
nvarchar (255) |
Hierarchy for Lithostratigraphy |
| _ChronostratigraphyPropertyHierarchyCache |
nvarchar (255) |
Hierarchy for Chronostratigraphy |
| _AverageAltitudeCache |
real |
Calculated average altitude as parsed from the location fields |
Depending on:
- Analysis
- CollectionAgent
- CollectionEvent
- CollectionEventLocalisation
- CollectionEventProperty
- CollectionSpecimen
- CollectionSpecimenID_UserAvailable
- CollectionSpecimenPart
- CollectionSpecimenProcessing
- CollectionSpecimenTransaction
- Identification
- IdentificationUnit
- IdentificationUnitAnalysis
- IdentificationUnitInPart
- Processing
- Transaction
Function FirstLinesSeries
Returns a table that lists all the collection event series with the first entries of related tables
| Parameter |
DataType |
Description |
| @CollectionSpecimenIDs |
varchar (8000) |
Comma separated list of CollectionSpecimenIDs for which the data should be retrieved |
| Column |
DataType |
Description |
| SeriesID |
int |
The ID of the related expedition. Relates to the PK of the table CollectionExpedition (foreign key). |
| SeriesParentID |
int |
The ID of the superior series of collection events |
| Description |
nvarchar (500) |
The description of the series of collection events as it will be printed on e.g. the label |
| SeriesCode |
nvarchar (50) |
The user defined code for a series of collection events |
| Geography |
geography (MAX) |
The geography of the series of collection events |
| Notes |
nvarchar (500) |
Notes on the collection event |
| DateStart |
datetime |
Point in time when the series of collection events started |
| DateEnd |
datetime |
Point in time when the series of collection events ended |
Depending on:
- CollectionEvent
- CollectionEventSeries
- CollectionSpecimen
- CollectionSpecimenID_UserAvailable
Function FirstLinesUnit_4
Returns a table that lists all the identification unit with the first entries of related tables
| Parameter |
DataType |
Description |
| @CollectionSpecimenIDs |
varchar (8000) |
Comma separated list of CollectionSpecimenIDs for which the data should be retrieved |
| @AnalysisIDs |
varchar (8000) |
Comma separated list of AnalysisIDs for which the data should be retrieved |
| @AnalysisStartDate |
date |
The start date of the range for analysis |
| @AnalysisEndDate |
date |
The end date of the range for analysis |
| Column |
DataType |
Description |
| IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
| CollectionSpecimenID |
int |
Refers to ID of CollectionEvent (= foreign key and part of primary key) |
| Accession_number |
nvarchar (50) |
Accession number of the specimen within the collection, e.g. “M-29834752” |
| Data_withholding_reason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
| Data_withholding_reason_for_collection_event |
nvarchar (255) |
If the data of the collection event are withhold, the reason for withholding the data, otherwise null |
| Data_withholding_reason_for_collector |
nvarchar (255) |
If the data of the collector are withhold, the reason for withholding the data, otherwise null |
| Collectors_event_number |
nvarchar (50) |
Number assigned to a collection event by the collector (= ‘field number’) |
| Collection_day |
tinyint |
The day of the date of the event or when the collection event started |
| Collection_month |
tinyint |
The month of the date of the event or when the collection event started |
| Collection_year |
smallint |
The year of the date of the event or when the collection event started |
| Collection_date_supplement |
nvarchar (100) |
Verbal or additional collection date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. The end date, if the collection event comprises a period. The time of the event, if necessary. |
| Collection_time |
varchar (50) |
The time of the event or when the collection event started |
| Collection_time_span |
varchar (50) |
The time span e.g. in seconds of the collection event |
| Country |
nvarchar (50) |
- |
| Locality_description |
nvarchar (255) |
Locality description of the locality exactly as written on the original label (i.e. without corrections during data entry) |
| Habitat_description |
nvarchar (255) |
Geo-ecological description of the locality exactly as written on the original label (i.e. without corrections during data entry) |
| Collecting_method |
nvarchar (255) |
Description of the method used for collecting the samples, e.g. traps, moist chambers, drag net |
| Collection_event_notes |
nvarchar (255) |
Notes about the collection event |
| Named_area |
nvarchar (255) |
Named area as e.g. derived from DiversityGazetteer corresponding to LocalisationSystemID = 7 |
| NamedAreaLocation2 |
nvarchar (255) |
Link to a named area as e.g. derived from DiversityGazetteer corresponding to LocalisationSystemID = 7 |
| Remove_link_to_gazetteer |
int |
Empty column reserved to remove a link to a named area as e.g. derived from DiversityGazetteer corresponding to LocalisationSystemID = 7 |
| Distance_to_location |
varchar (50) |
Distance from the specified place to the real location of the collection site (m) |
| Direction_to_location |
varchar (50) |
Direction from the specified place to the real location of the collection site (Degrees rel. to north) |
| Longitude |
nvarchar (255) |
Longitude derived from WGS84 corresponding to LocalisationSystemID = 8 |
| Latitude |
nvarchar (255) |
Latitude derived from WGS84 corresponding to LocalisationSystemID = 8 |
| Coordinates_accuracy |
nvarchar (50) |
Accuracy of coordinates derived from WGS84 corresponding to LocalisationSystemID = 8 |
| Link_to_GoogleMaps |
int |
Empty column reserved for retrieval of WGS84 coordinates via e.g. GoogleMaps corresponding to LocalisationSystemID = 8 |
| Altitude_from |
nvarchar (255) |
Lower value of altitude range corresponding to LocalisationSystemID = 4 |
| Altitude_to |
nvarchar (255) |
Upper value of altitude range corresponding to LocalisationSystemID = 4 |
| Altitude_accuracy |
nvarchar (50) |
Accuracy of altitude range corresponding to LocalisationSystemID = 4 |
| Notes_for_Altitude |
nvarchar (255) |
Notes for altitude range corresponding to LocalisationSystemID = 4 |
| MTB |
nvarchar (255) |
TK25 corresponding to LocalisationSystemID = 3 |
| Quadrant |
nvarchar (255) |
Quadrant of TK25 corresponding to LocalisationSystemID = 3 |
| Notes_for_MTB |
nvarchar (255) |
Notes for TK25 corresponding to LocalisationSystemID = 3 |
| Sampling_plot |
nvarchar (255) |
Name of the sampling plot corresponding to LocalisationSystemID = 13 |
| Link_to_SamplingPlots |
nvarchar (255) |
Link to the sampling plot corresponding to LocalisationSystemID = 13 |
| Remove_link_to_SamplingPlots |
int |
Empty column reserverd to remove link to the sampling plot corresponding to LocalisationSystemID = 13 |
| Accuracy_of_sampling_plot |
nvarchar (50) |
Accuracy of the sampling plot coordinates corresponding to LocalisationSystemID = 13 |
| Latitude_of_sampling_plot |
real |
Latitude of the sampling plot coordinates corresponding to LocalisationSystemID = 13 |
| Longitude_of_sampling_plot |
real |
Longitude of the sampling plot coordinates corresponding to LocalisationSystemID = 13 |
| Geographic_region |
nvarchar (255) |
Geographic region corresponding to LocalisationSystemID = 10 |
| Lithostratigraphy |
nvarchar (255) |
Lithostratigraphy corresponding to LocalisationSystemID = 30 |
| Chronostratigraphy |
nvarchar (255) |
Chronostratigraphy corresponding to LocalisationSystemID = 20 |
| Collectors_name |
nvarchar (255) |
Name of the Collector |
| Link_to_DiversityAgents |
varchar (255) |
Link for the first collector to DiversityAgents |
| Remove_link_for_collector |
int |
Empty column reserved for removal of ink for the first collector |
| Collectors_number |
nvarchar (50) |
Number assigned to a specimen or a batch of specimens by the collector during the collection event (= ‘field number’) |
| Notes_about_collector |
nvarchar (MAX) |
Notes about the first collector |
| Accession_day |
tinyint |
The day of the date when the specimen was acquired in the collection |
| Accession_month |
tinyint |
The month of the date when the specimen was acquired in the collection |
| Accession_year |
smallint |
The year of the date when the specimen was acquired in the collection |
| Accession_date_supplement |
nvarchar (255) |
Verbal or additional accession date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
| Depositors_name |
nvarchar (255) |
The name of the depositor(s) (person or organization responsible for deposition). Where entire collections are deposited, this should also contain the collection name (e.g. ‘Herbarium P. Döbbler’) |
| Depositors_link_to_DiversityAgents |
varchar (255) |
The link for the depositor e.g. to DiversityAgents |
| Remove_link_for_Depositor |
int |
Empty column reserved to remove the link for the depositor e.g. to DiversityAgents |
| Depositors_accession_number |
nvarchar (50) |
Accession number of the specimen within the previous or original collection, e.g. ‘D-23948’ |
| Exsiccata_abbreviation |
nvarchar (255) |
If specimen is an exsiccata: Standard abbreviation of the exsiccata (not necessarily a unique identifier; editors or publication places may change over time) |
| Link_to_DiversityExsiccatae |
varchar (255) |
Link to DiversityExsiccatae |
| Remove_link_to_exsiccatae |
int |
Empty column reserved to remove the link to DiversityExsiccatae |
| Exsiccata_number |
nvarchar (50) |
If specimen is an exsiccata: Number of current specimen within the exsiccata series |
| Original_notes |
nvarchar (MAX) |
Notes found on the label of the specimen by the original collector or from a later revision |
| Additional_notes |
nvarchar (MAX) |
Additional notes made by the editor of the specimen record, e.g. ‘doubtful identification/locality’ |
| Internal_notes |
nvarchar (MAX) |
Internal notes which should not be published e.g. on websites |
| Label_title |
nvarchar (255) |
The title of the label e.g. for printing labels. |
| Label_type |
nvarchar (50) |
Printed, typewritten, typewritten with handwriting added, entirely in handwriting, etc. |
| Label_transcription_state |
nvarchar (50) |
The state of the transcription of a label into the database: ‘Not started’, ‘incomplete’, ‘complete’ |
| Label_transcription_notes |
nvarchar (255) |
User defined notes on the transcription of the label into the database |
| Problems |
nvarchar (255) |
Description of a problem which occurred during data editing. Typically these entries should be deleted after help has been obtained. Do not enter scientific problems here; use AdditionalNotes for such permanent problems! |
| Taxonomic_group |
nvarchar (50) |
Taxonomic group the organism, identified by this unit, belongs to. Groups listed in table CollTaxonomicGroup_Enum (= foreign key) |
| Relation_type |
nvarchar (50) |
The relation of a unit to its substrate, e.g. parasitism, symbiosis etc. as stored in CollRelationType_Enum (= foreign key) |
| Colonised_substrate_part |
nvarchar (255) |
If a substrate association exists: part of the substrate which is affected in the interaction (e.g. ’leaves’, if a fungus is growing on the leaves of an infected plant) |
| Related_organism |
nvarchar (200) |
Last identification of the related organism |
| Life_stage |
nvarchar (255) |
Examples: ‘II, III’ for spore generations of rusts or ‘seed’, ‘seedling’ etc. for higher plants |
| Gender |
nvarchar (50) |
The sex of the organism, e.g. ‘female’ |
| Number_of_units |
smallint |
The number of units of this organism, e.g. 400 beetles in a bottle |
| Circumstances |
nvarchar (50) |
Circumstances of the occurence of the organism |
| Order_of_taxon |
nvarchar (255) |
Order of the first taxon |
| Family_of_taxon |
nvarchar (255) |
Family of the first taxon |
| Identifier_of_organism |
nvarchar (50) |
Identifier of the first organism |
| Description_of_organism |
nvarchar (50) |
Description of the first organism |
| Only_observed |
bit |
True, if the organism was only observed rather than collected. It is therefore not present on the preserved specimen. Example: Tree under which the collected mycorrhizal fungus grew. |
| Notes_for_organism |
nvarchar (MAX) |
Notes for the first organism |
| Taxonomic_name |
nvarchar (255) |
Valid name of the species (including the taxonomic author where available). Example: ‘Rosa canina L.’ |
| Link_to_DiversityTaxonNames |
varchar (255) |
Link for first identification of first organism to DiversityTaxonNames |
| Remove_link_for_identification |
int |
Empty column to remove link for first identification of first organism to DiversityTaxonNames |
| Vernacular_term |
nvarchar (255) |
Name or term other than a taxonomic (= scientific) name, e.g. ‘pine’, ’limestone’, ‘conifer’, ‘hardwood’ |
| Identification_day |
tinyint |
The day of the identification |
| Identification_month |
tinyint |
The month of the identification |
| Identification_year |
smallint |
The year of the identification. The year may be empty if only the day or month are known. |
| Identification_category |
nvarchar (50) |
Category of the identification e.g. ‘determination’, ‘confirmation’, ‘absence’ (= foreign key, see table CollIdentificationCategory_Enum) |
| Identification_qualifier |
nvarchar (50) |
Qualification of the identification e.g. “cf.”," aff.", “sp. nov.” (= foreign key, see table CollIdentificationQualifier_Enum) |
| Type_status |
nvarchar (50) |
If identification unit is type of a taxonomic name: holotype, syntype, etc. (= foreign key, see table CollTypeStatus_Enum) |
| Type_notes |
nvarchar (MAX) |
Notes on the typification of this specimen |
| Notes_for_identification |
nvarchar (MAX) |
Notes for first identification of first organism to DiversityTaxonNames |
| Determiner |
nvarchar (255) |
Agent responsible for first identification of first organism |
| Link_to_DiversityAgents_for_determiner |
varchar (255) |
Agent for responsible for first identification of first organism to e.g. DiversityAgents |
| Remove_link_for_determiner |
int |
Empty column reservet to remove link for agent for responsible for first identification of first organism to e.g. DiversityAgents |
| Analysis_0 |
nvarchar (50) |
Name of analysis 0 according to list of AnalysisIDs |
| AnalysisID_0 |
int |
ID of analysis 0 according to list of AnalysisIDs |
| Analysis_number_0 |
nvarchar (50) |
Number of analysis 0 according to list of AnalysisIDs |
| Analysis_result_0 |
nvarchar (MAX) |
Result of analysis 0 according to list of AnalysisIDs |
| Analysis_1 |
nvarchar (50) |
Name of analysis_1 according to list of AnalysisIDs |
| AnalysisID_1 |
int |
ID of analysis_1 according to list of AnalysisIDs |
| Analysis_number_1 |
nvarchar (50) |
Number of analysis_1 according to list of AnalysisIDs |
| Analysis_result_1 |
nvarchar (MAX) |
Result of analysis_1 according to list of AnalysisIDs |
| Analysis_2 |
nvarchar (50) |
Name of analysis_2 according to list of AnalysisIDs |
| AnalysisID_2 |
int |
ID of analysis_2 according to list of AnalysisIDs |
| Analysis_number_2 |
nvarchar (50) |
Number of analysis_2 according to list of AnalysisIDs |
| Analysis_result_2 |
nvarchar (MAX) |
Result of analysis_2 according to list of AnalysisIDs |
| Analysis_3 |
nvarchar (50) |
Name of analysis_3 according to list of AnalysisIDs |
| AnalysisID_3 |
int |
ID of analysis_3 according to list of AnalysisIDs |
| Analysis_number_3 |
nvarchar (50) |
Number of analysis_3 according to list of AnalysisIDs |
| Analysis_result_3 |
nvarchar (MAX) |
Result of analysis_3 according to list of AnalysisIDs |
| Analysis_4 |
nvarchar (50) |
Name of analysis_4 according to list of AnalysisIDs |
| AnalysisID_4 |
int |
ID of analysis_4 according to list of AnalysisIDs |
| Analysis_number_4 |
nvarchar (50) |
Number of analysis_4 according to list of AnalysisIDs |
| Analysis_result_4 |
nvarchar (MAX) |
Result of analysis_4 according to list of AnalysisIDs |
| Analysis_5 |
nvarchar (50) |
Name of analysis_5 according to list of AnalysisIDs |
| AnalysisID_5 |
int |
ID of analysis_5 according to list of AnalysisIDs |
| Analysis_number_5 |
nvarchar (50) |
Number of analysis_5 according to list of AnalysisIDs |
| Analysis_result_5 |
nvarchar (MAX) |
Result of analysis_5 according to list of AnalysisIDs |
| Analysis_6 |
nvarchar (50) |
Name of analysis_6 according to list of AnalysisIDs |
| AnalysisID_6 |
int |
ID of analysis_6 according to list of AnalysisIDs |
| Analysis_number_6 |
nvarchar (50) |
Number of analysis_6 according to list of AnalysisIDs |
| Analysis_result_6 |
nvarchar (MAX) |
Result of analysis_6 according to list of AnalysisIDs |
| Analysis_7 |
nvarchar (50) |
Name of analysis_7 according to list of AnalysisIDs |
| AnalysisID_7 |
int |
ID of analysis_7 according to list of AnalysisIDs |
| Analysis_number_7 |
nvarchar (50) |
Number of analysis_7 according to list of AnalysisIDs |
| Analysis_result_7 |
nvarchar (MAX) |
Result of analysis_7 according to list of AnalysisIDs |
| Analysis_8 |
nvarchar (50) |
Name of analysis_8 according to list of AnalysisIDs |
| AnalysisID_8 |
int |
ID of analysis_8 according to list of AnalysisIDs |
| Analysis_number_8 |
nvarchar (50) |
Number of analysis_8 according to list of AnalysisIDs |
| Analysis_result_8 |
nvarchar (MAX) |
Result of analysis_8 according to list of AnalysisIDs |
| Analysis_9 |
nvarchar (50) |
Name of analysis_9 according to list of AnalysisIDs |
| AnalysisID_9 |
int |
ID of analysis_9 according to list of AnalysisIDs |
| Analysis_number_9 |
nvarchar (50) |
Number of analysis_9 according to list of AnalysisIDs |
| Analysis_result_9 |
nvarchar (MAX) |
Result of analysis_9 according to list of AnalysisIDs |
| Collection |
int |
- |
| Material_category |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. (= foreign key, see table CollMaterialCategory_Enum) |
| Storage_location |
nvarchar (255) |
A code identifying the place where the specimen is stored within the collection. Frequently the accepted scientific name is used as storage location code. |
| Stock |
float |
Number of stock units, if the specimen is stored in separated units e.g. several boxes or vessels (max. 255) |
| Preparation_method |
nvarchar (MAX) |
The method used for the preparation of the part of the specimen, e.g. the inoculation method for cultures |
| Preparation_date |
datetime |
Point in time when the part was preparated e.g when it was separated from the source object |
| Notes_for_part |
nvarchar (MAX) |
Notes for specimen part |
| _TransactionID |
int |
Unique ID for the table Transaction (= foreign key and part of primary key) |
| _Transaction |
nvarchar (200) |
Title of first transaction |
| On_loan |
int |
If part is on loan according to first transaction |
| _CollectionEventID |
int |
Unique ID for the table CollectionEvent (= primary key) |
| _IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
| _IdentificationSequence |
smallint |
The sequence of the identifications. The last identification (having the highest sequence) is regarded as valid |
| _SpecimenPartID |
int |
Unique ID of the part of the collection specimen (= part of primary key). |
| _CoordinatesAverageLatitudeCache |
real |
Average Latitude of WGS84 Coordinates |
| _CoordinatesAverageLongitudeCache |
real |
Average Longitude of WGS84 Coordinates |
| _CoordinatesLocationNotes |
nvarchar (255) |
Notes for WGS84 Coordinates |
| _GeographicRegionPropertyURI |
varchar (255) |
URI of Geographic Region |
| _LithostratigraphyPropertyURI |
varchar (255) |
URI for Lithostratigraphy |
| _ChronostratigraphyPropertyURI |
varchar (255) |
URI for Chronostratigraphy |
| _NamedAverageLatitudeCache |
real |
Average Latitude for Named Area |
| _NamedAverageLongitudeCache |
real |
Average Longitude for Named Area |
| _LithostratigraphyPropertyHierarchyCache |
nvarchar (255) |
Hierarchy for Lithostratigraphy |
| _ChronostratigraphyPropertyHierarchyCache |
nvarchar (255) |
Hierarchy for Chronostratigraphy |
| _AverageAltitudeCache |
real |
Average Altitude |
Depending on:
- Analysis
- CollectionAgent
- CollectionEvent
- CollectionEventLocalisation
- CollectionEventProperty
- CollectionSpecimen
- CollectionSpecimenID_UserAvailable
- CollectionSpecimenPart
- CollectionSpecimenTransaction
- Identification
- IdentificationUnit
- IdentificationUnitAnalysis
- Transaction
Function LocalisationSystem_List
| Parameter |
DataType |
Description |
| @LanguageCode |
char (2) |
- |
| Column |
DataType |
Description |
| LocalisationSystemID |
int |
Unique ID for the localisation system (= Primary key) |
| LocalisationSystemParentID |
int |
LocalisationSystemID of the superior LocalisationSystem |
| LocalisationSystemName |
nvarchar (50) |
Name of the system used for the determination of the place of the collection, e. g. Gauss-Krüger, MTB, GIS |
| MeasurementUnit |
nvarchar (100) |
- |
| DefaultAccuracyOfLocalisation |
nvarchar (100) |
The default for the accuracy of values which can be reached with this method |
| DiversityModule |
nvarchar (50) |
- |
| ParsingMethod |
nvarchar (500) |
- |
| LocalisationSystemType |
nvarchar (500) |
- |
| Description |
nvarchar (255) |
Description of the localisation method |
| Abbreviation |
nvarchar (50) |
- |
| DescriptionLocation1 |
nvarchar (255) |
Description of the attribute Location1 in the table CollectionGeography as displayed in the user interface |
| AbbreviationLocation1 |
nvarchar (50) |
- |
| DescriptionLocation2 |
nvarchar (255) |
Description of the attribute Location2 in the table CollectionGeography as displayed in the user interface |
| AbbreviationLocation2 |
nvarchar (50) |
- |
Function ManagerCollectionList
Returns a table that lists all the collections a Manager has access to, including the child collections.
| Column |
DataType |
Description |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
| CollectionName |
nvarchar (255) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
| CollectionAcronym |
nvarchar (50) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
| AdministrativeContactAgentURI |
nvarchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
| Description |
nvarchar (4000) |
A short description of the collection |
| Location |
nvarchar (1000) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
| DisplayOrder |
varchar (255) |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
Depending on:
- Collection
- CollectionChildNodes
- CollectionManager
Function MethodChildNodes
All child nodes of a given method related via the MethodParentID
| Parameter |
DataType |
Description |
| @ID |
int |
MethodID for which the depending methods should be returned |
| Column |
DataType |
Description |
| MethodID |
int |
ID of the Method (Primary key) |
| MethodParentID |
int |
MethodID of the parent Method, if it belongs to a certain type documented in this table |
| DisplayText |
nvarchar (50) |
Name of the Method as e.g. shown in user interface |
| Description |
nvarchar (500) |
Description of the Method |
| ForCollectionEvent |
bit |
If a method may be used during a collection event |
| Notes |
nvarchar (1000) |
Notes on this method |
| MethodURI |
varchar (255) |
URI referring to an external documentation of the Method |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries |
| RowGUID |
uniqueidentifier |
- |
Depending on:
Function MethodHierarchy
The hierarchy of a given method
| Parameter |
DataType |
Description |
| @MethodID |
int |
ID of the Method (Primary key) |
| Column |
DataType |
Description |
| MethodID |
int |
ID of the Method (Primary key) |
| MethodParentID |
int |
MethodID of the parent Method, if it belongs to a certain type documented in this table |
| DisplayText |
nvarchar (50) |
Name of the Method as e.g. shown in user interface |
| Description |
nvarchar (500) |
Description of the Method |
| Notes |
nvarchar (1000) |
Notes on this method |
| MethodURI |
varchar (255) |
URI referring to an external documentation of the Method |
| ForCollectionEvent |
bit |
If a method may be used during a collection event |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries |
Depending on:
Function MethodHierarchyAll
All methods including a column displaying the hierarchy of the method
| Column |
DataType |
Description |
| MethodID |
int |
ID of the Method (Primary key) |
| MethodParentID |
int |
MethodID of the parent Method, if it belongs to a certain type documented in this table |
| DisplayText |
nvarchar (255) |
Name of the Method as e.g. shown in user interface |
| Description |
nvarchar (MAX) |
Description of the Method |
| Notes |
nvarchar (MAX) |
Notes on this method |
| MethodURI |
varchar (255) |
URI referring to an external documentation of the Method |
| ForCollectionEvent |
bit |
If a method may be used during a collection event |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries |
| HierarchyDisplayText |
nvarchar (900) |
Hierarchy of the method separated by |
Depending on:
Function NameList
| Parameter |
DataType |
Description |
| @NameID |
int |
- |
| @NameStart |
nvarchar (50) |
- |
| Column |
DataType |
Description |
| NameID |
int |
- |
| TaxonName |
nvarchar (255) |
- |
Function NameListMyxomycetes
| Parameter |
DataType |
Description |
| @NameID |
int |
- |
| @NameStart |
nvarchar (50) |
- |
| Column |
DataType |
Description |
| NameID |
int |
- |
| TaxonName |
nvarchar (200) |
- |
Function NameListPlants
| Parameter |
DataType |
Description |
| @NameID |
int |
- |
| @NameStart |
nvarchar (50) |
- |
| Column |
DataType |
Description |
| NameID |
int |
- |
| TaxonName |
nvarchar (200) |
- |
Function NextFreeAccNr
returns next free accession number similar to given parameter. Assumes that accession numbers have a pattern like M-0023423 or HAL 25345 or GLM3453 with a leading string and a numeric end
DataType: nvarchar (50)
| Parameter |
DataType |
Description |
| @AccessionNumber |
nvarchar (50) |
Accession number of the specimen within the collection, e.g. “M-29834752” |
Depending on:
Function NextFreeAccNumber
The next free accession number starting like parameter 1, optional inclusion of specimen (parameter 2) and parts (parameter 3)
DataType: nvarchar (50)
| Parameter |
DataType |
Description |
| @AccessionNumber |
nvarchar (50) |
Accession number of the specimen within the collection, e.g. “M-29834752” |
| @IncludeSpecimen |
bit |
If specimen should be included in search |
| @IncludePart |
bit |
If parts should be included in search |
Depending on:
- CollectionSpecimen
- CollectionSpecimenPart
Function PrivacyConsentInfo
Provides a link to common information about the DiversityWorkbench
DataType: varchar (900)
Function ProcessingChildNodes
Returns a result set that lists all the processings within a hierarchy starting at the topmost processing related to the given item
| Parameter |
DataType |
Description |
| @ID |
int |
ID of the parent processing |
| Column |
DataType |
Description |
| ProcessingID |
int |
ID of the processing (primary key) |
| ProcessingParentID |
int |
The ID of the superior type of the processing |
| DisplayText |
nvarchar (50) |
The display text of the processing as shown e.g. in a user interface |
| Description |
nvarchar (500) |
Description of the processing |
| Notes |
nvarchar (1000) |
Notes on the processing |
| ProcessingURI |
varchar (255) |
A URI for a processing as defined in an external data source |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries |
| RowGUID |
uniqueidentifier |
- |
Depending on:
- Processing
- ProcessingChildNodes
Function ProcessingHierarchy
Returns a table that lists all the analysis items related to the given processing.
| Parameter |
DataType |
Description |
| @ID |
int |
ID of the parent processing |
| Column |
DataType |
Description |
| ProcessingID |
int |
ID of the processing (primary key) |
| ProcessingParentID |
int |
The ID of the superior type of the processing |
| DisplayText |
nvarchar (50) |
The display text of the processing as shown e.g. in a user interface |
| Description |
nvarchar (500) |
Description of the processing |
| Notes |
nvarchar (1000) |
Notes on the processing |
| ProcessingURI |
varchar (255) |
A URI for a processing as defined in an external data source |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries |
| RowGUID |
uniqueidentifier |
- |
Depending on:
- Processing
- ProcessingChildNodes
Function ProcessingHierarchyAll
Returns a table that lists all the Processings including their hierarchy
| Column |
DataType |
Description |
| ProcessingID |
int |
ID of the processing (primary key) |
| ProcessingParentID |
int |
The ID of the superior type of the processing |
| DisplayText |
nvarchar (50) |
The display text of the processing as shown e.g. in a user interface |
| Description |
nvarchar (MAX) |
Description of the processing |
| Notes |
nvarchar (MAX) |
Notes on the processing |
| ProcessingURI |
varchar (255) |
A URI for a processing as defined in an external data source |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries |
| HierarchyDisplayText |
varchar (900) |
Hierarchy of the processings as DisplayText separated by |
Depending on:
Function ProcessingListForPart
Returns a table that lists all the processing items related to the given part.Tthe list depends upon the processing types available for a material category and the projects available for a processing
| Parameter |
DataType |
Description |
| @CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
| @SpecimenPartID |
int |
Unique ID of the part of the collection specimen (= part of primary key). |
| Column |
DataType |
Description |
| ProcessingID |
int |
ID of the processing (primary key) |
| ProcessingParentID |
int |
The ID of the superior type of the processing |
| DisplayText |
nvarchar (50) |
The display text of the processing as shown e.g. in a user interface |
| Description |
nvarchar (MAX) |
Description of the processing |
| Notes |
nvarchar (MAX) |
Notes on the storage of the sample |
| ProcessingURI |
varchar (255) |
A URI for a processing as defined in an external data source |
| DisplayTextHierarchy |
nvarchar (255) |
Hierarchy of the processings as DisplayText separated by |
Depending on:
- CollectionProject
- CollectionSpecimen
- CollectionSpecimenPart
- Processing
- ProcessingChildNodes
- ProcessingHierarchy
- ProcessingMaterialCategory
- ProjectProcessing
Function ProcessingProjectList
Returns a table that lists all the Processing items related to the given project.
| Parameter |
DataType |
Description |
| @ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
| Column |
DataType |
Description |
| ProcessingID |
int |
ID of the processing (primary key) |
| ProcessingParentID |
int |
The ID of the superior type of the processing |
| DisplayText |
nvarchar (50) |
The display text of the processing as shown e.g. in a user interface |
| Description |
nvarchar (500) |
Description of the processing |
| Notes |
nvarchar (1000) |
Notes on the processing |
| ProcessingURI |
varchar (255) |
A URI for a processing as defined in an external data source |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries |
| RowGUID |
uniqueidentifier |
- |
Depending on:
- Processing
- ProcessingChildNodes
- ProjectProcessing
Function ProjectDataLastChanges
retrieval of the last update in data of a project
DataType: datetime
| Parameter |
DataType |
Description |
| @ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
Depending on:
- CollectionEvent
- CollectionProject
- CollectionProject_log
- CollectionSpecimen
Function RequesterCollectionList
Returns a table that lists all the collections a requester has access to, including the child collections if allowed
| Column |
DataType |
Description |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
| CollectionName |
nvarchar (50) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
| CollectionAcronym |
nvarchar (50) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
| AdministrativeContactAgentURI |
nvarchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
| Description |
nvarchar (500) |
A short description of the collection |
| Location |
nvarchar (1000) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
| DisplayOrder |
varchar (255) |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
Depending on:
- Collection
- CollectionHierarchy
- CollectionRequester
Function StableIdentifier
Returns a stable identfier for a dataset. Relies on an entry in ProjectProxy
DataType: varchar (500)
| Parameter |
DataType |
Description |
| @ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
| @CollectionSpecimenID |
int |
CollectionSpecimenID that should be included in the StableIdentifier |
| @IdentificationUnitID |
int |
IdentificationUnitID that should be included in the StableIdentifier |
| @SpecimenPartID |
int |
SpecimenPartID that should be included in the StableIdentifier |
Depending on:
Function TaskChildNodes
All child nodes of a given Task related via the TaskParentID
| Parameter |
DataType |
Description |
| @ID |
int |
TaskParentID for which all child nodes of a given Task should be returned |
| Column |
DataType |
Description |
| TaskID |
int |
ID of the Task (primary key) |
| TaskParentID |
int |
The ID of the superior type of the Task |
| DisplayText |
nvarchar (50) |
The display text of the Task as shown e.g. in a user interface |
| Description |
nvarchar (500) |
Description of the Task |
| Notes |
nvarchar (1000) |
Notes on the Task |
| TaskURI |
varchar (255) |
A URI for a Task as defined in an external data source |
| Type |
varchar (50) |
The type of the task as defined in table TaskType_Enum |
| ModuleTitle |
varchar (50) |
The title for module related data for collection tasks. Not available in depending collection task if empty |
| ModuleType |
varchar (50) |
The DiversityWorkbench module to which a task is related. Not available in depending collection task if empty |
| SpecimenPartType |
nvarchar (50) |
The description of the collection specimen part to which a task is related. Not available in depending collection task if empty |
| TransactionType |
nvarchar (50) |
The description of the transaction to which a task is related. Not available in depending collection task if empty |
| ResultType |
varchar (50) |
The display text for the results as shown in a user interface. Not available in depending collection task if empty |
| DateType |
varchar (50) |
The date and time details defined for a task. Not available in depending collection task if empty |
| DateBeginType |
nvarchar (50) |
The definition of the begin for date and time details defined for a task. Not available in depending collection task if empty |
| DateEndType |
nvarchar (50) |
The definition of the end for date and time details defined for a task. Not available in depending collection task if empty |
| NumberType |
varchar (50) |
The definition for the numeric value as shown in a user interface. Not available in depending collection task if empty |
| BoolType |
varchar (50) |
The definition for the boolean value as shown in a user interface. Not available in depending collection task if empty |
| MetricType |
nvarchar (50) |
The definition for the metric as shown in a user interface. Not available in depending collection task if empty |
| MetricUnit |
nvarchar (50) |
The unit of the metric, e.g. °C for temperature |
| ResponsibleType |
nvarchar (50) |
The definition for the responsible agent as shown in a user interface. Not available in depending collection task if empty |
| DescriptionType |
varchar (50) |
The definition for the description as shown in a user interface. Not available in depending collection task if empty |
| NotesType |
varchar (50) |
The definition for the notes as shown in a user interface. Not available in depending collection task if empty |
| UriType |
varchar (50) |
The definition for the URI as shown in a user interface. Not available in depending collection task if empty |
| RowGUID |
uniqueidentifier |
- |
Depending on:
Function TaskCollectionHierarchySeparator
Returns a separator used between collection and task hierarchy
DataType: nchar (3)
Function TaskHierarchy
The hierarchy of a given Task
| Parameter |
DataType |
Description |
| @TaskID |
int |
ID of the Task (primary key) |
| Column |
DataType |
Description |
| TaskID |
int |
ID of the Task (primary key) |
| TaskParentID |
int |
The ID of the superior type of the Task |
| DisplayText |
nvarchar (50) |
The display text of the Task as shown e.g. in a user interface |
| Description |
nvarchar (500) |
Description of the Task |
| Notes |
nvarchar (1000) |
Notes on the Task |
| TaskURI |
varchar (255) |
A URI for a Task as defined in an external data source |
| Type |
varchar (50) |
The type of the task as defined in table TaskType_Enum |
| ModuleTitle |
varchar (50) |
The title for module related data for collection tasks. Not available in depending collection task if empty |
| ModuleType |
varchar (50) |
The DiversityWorkbench module to which a task is related. Not available in depending collection task if empty |
| SpecimenPartType |
nvarchar (50) |
The description of the collection specimen part to which a task is related. Not available in depending collection task if empty |
| TransactionType |
nvarchar (50) |
The description of the transaction to which a task is related. Not available in depending collection task if empty |
| ResultType |
varchar (50) |
The display text for the results as shown in a user interface. Not available in depending collection task if empty |
| DateType |
varchar (50) |
The date and time details defined for a task. Not available in depending collection task if empty |
| DateBeginType |
nvarchar (50) |
The definition of the begin for date and time details defined for a task. Not available in depending collection task if empty |
| DateEndType |
nvarchar (50) |
The definition of the end for date and time details defined for a task. Not available in depending collection task if empty |
| NumberType |
varchar (50) |
The definition for the numeric value as shown in a user interface. Not available in depending collection task if empty |
| BoolType |
varchar (50) |
The definition for the boolean value as shown in a user interface. Not available in depending collection task if empty |
| MetricType |
nvarchar (50) |
The definition for the metric as shown in a user interface. Not available in depending collection task if empty |
| MetricUnit |
nvarchar (50) |
The unit of the metric, e.g. °C for temperature |
| ResponsibleType |
nvarchar (50) |
The definition for the responsible agent as shown in a user interface. Not available in depending collection task if empty |
| DescriptionType |
varchar (50) |
The definition for the description as shown in a user interface. Not available in depending collection task if empty |
| NotesType |
varchar (50) |
The definition for the notes as shown in a user interface. Not available in depending collection task if empty |
| UriType |
varchar (50) |
The definition for the URI as shown in a user interface. Not available in depending collection task if empty |
Depending on:
Function TaskHierarchyAll
All Tasks including a column displaying the hierarchy of the Task
| Column |
DataType |
Description |
| TaskID |
int |
ID of the Task (primary key) |
| TaskParentID |
int |
The ID of the superior type of the Task |
| DisplayText |
nvarchar (50) |
The display text of the Task as shown e.g. in a user interface |
| Description |
nvarchar (MAX) |
Description of the Task |
| Notes |
nvarchar (MAX) |
Notes on the Task |
| TaskURI |
varchar (255) |
A URI for a Task as defined in an external data source |
| Type |
varchar (50) |
The type of the task as defined in table TaskType_Enum |
| ModuleTitle |
varchar (50) |
The title for module related data for collection tasks. Not available in depending collection task if empty |
| ModuleType |
varchar (50) |
The DiversityWorkbench module to which a task is related. Not available in depending collection task if empty |
| SpecimenPartType |
nvarchar (50) |
The description of the collection specimen part to which a task is related. Not available in depending collection task if empty |
| TransactionType |
nvarchar (50) |
The description of the transaction to which a task is related. Not available in depending collection task if empty |
| ResultType |
varchar (50) |
The display text for the results as shown in a user interface. Not available in depending collection task if empty |
| DateType |
varchar (50) |
The date and time details defined for a task. Not available in depending collection task if empty |
| DateBeginType |
nvarchar (50) |
The definition of the begin for date and time details defined for a task. Not available in depending collection task if empty |
| DateEndType |
nvarchar (50) |
The definition of the end for date and time details defined for a task. Not available in depending collection task if empty |
| NumberType |
varchar (50) |
The definition for the numeric value as shown in a user interface. Not available in depending collection task if empty |
| BoolType |
varchar (50) |
The definition for the boolean value as shown in a user interface. Not available in depending collection task if empty |
| MetricType |
nvarchar (50) |
The definition for the metric as shown in a user interface. Not available in depending collection task if empty |
| MetricUnit |
nvarchar (50) |
The unit of the metric, e.g. °C for temperature |
| ResponsibleType |
nvarchar (50) |
The definition for the responsible agent as shown in a user interface. Not available in depending collection task if empty |
| DescriptionType |
varchar (50) |
The definition for the description as shown in a user interface. Not available in depending collection task if empty |
| NotesType |
varchar (50) |
The definition for the notes as shown in a user interface. Not available in depending collection task if empty |
| UriType |
varchar (50) |
The definition for the URI as shown in a user interface. Not available in depending collection task if empty |
| HierarchyDisplayText |
nvarchar (900) |
Hierarchy of the task starting at top Task separated by string defined in dbo.TaskHierarchySeparator() |
| HierarchyDisplayTextInvers |
nvarchar (900) |
Hierarchy of the task ending at top Task separated by string defined in dbo.TaskHierarchySeparator() |
Depending on:
- Task
- TaskHierarchySeparator
Function TaskHierarchySeparator
Returns a separator used within a task hierarchy
DataType: nchar (3)
Function TaxonWithQualifier
Generates a valid name for a taxon using the taxonomic name and the identification qualifier
DataType: nvarchar (500)
| Parameter |
DataType |
Description |
| @Taxon |
nvarchar (500) |
Taxonomic name used as base for transformation |
| @Qualifier |
nvarchar (50) |
Qualifier used as base for transformation |
Function TransactionChildNodes
Returns a result set that lists all the transactions within a hierarchy starting at the topmost transaction related to the given transaction.
| Parameter |
DataType |
Description |
| @ID |
int |
TransactionID of the topmost transaction within the hierarchy that should be returned |
| Column |
DataType |
Description |
| TransactionID |
int |
Unique ID for the transaction (= primary key) |
| ParentTransactionID |
int |
The ID of a preceeding transaction of a superior transaction, if transactions are organized in a hierarchy |
| TransactionType |
nvarchar (50) |
Type of the transaction, e.g. gift in or out, exchange in or out, purchase in or out |
| TransactionTitle |
nvarchar (200) |
The title of the transaction as e.g. shown in an user interface |
| ReportingCategory |
nvarchar (50) |
A group defined for the transaction, e.g. a taxonomic group as used for exchange balancing |
| AdministratingCollectionID |
int |
ID of the collection which is responsible for the administration of the transaction. |
| MaterialDescription |
nvarchar (MAX) |
Description of the material of this transaction |
| MaterialSource |
nvarchar (500) |
The source of the material within a transaction, e.g. a excavation |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. |
| MaterialCollectors |
nvarchar (MAX) |
The collectors of the material |
| FromCollectionID |
int |
The ID of the collection from which the specimen were transfered, e.g. the donating collection of a gift |
| FromTransactionPartnerName |
nvarchar (255) |
Name of the person or institution from which the specimen were transfered, e.g. the donator of a gift |
| FromTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
| FromTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the source of the specimen, e.g. the donating collection of a gift |
| ToCollectionID |
int |
The ID of the collection to which the specimen were transfered, e.g. the receiver of a gift |
| ToTransactionPartnerName |
nvarchar (255) |
Name of the person or institution to which the specimen were transfered, e.g. the receiver of a gift |
| ToTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
| ToTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the destination of the specimen, e.g. the receiving collection of a gift |
| ToRecipient |
nvarchar (255) |
The recipient receiving the transaction e.g. if not derived from the link to DiversityAgents |
| NumberOfUnits |
int |
The number of units which were (initially) included in the transaction |
| Investigator |
nvarchar (200) |
The investigator for whose study a transacted material was sent |
| TransactionComment |
nvarchar (MAX) |
Comments on the exchanged material addressed to the transaction partner |
| BeginDate |
datetime |
Date when the transaction started |
| AgreedEndDate |
datetime |
End of the transaction period, e.g. if the time for borrowing the specimen is restricted |
| ActualEndDate |
datetime |
Actual end of the transaction after a prolonation when e.g. the date of return for a loan was prolonged by the owner |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction not to be published e.g. on a web page |
| ResponsibleName |
nvarchar (255) |
The person responsible for this transaction |
| ResponsibleAgentURI |
varchar (255) |
The URI of the person, team or organisation responsible for the data (see e.g. module DiversityAgents) |
| DateSupplement |
nvarchar (100) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. |
Depending on:
Function TransactionChildNodesAccess
Returns a result set that lists all the items within a hierarchy starting at the topmost item related to the given item where the current user has no access according to the restriction in TransactionList.
| Parameter |
DataType |
Description |
| @ID |
int |
TransactionID of the topmost transaction within the hierarchy that should be returned |
| Column |
DataType |
Description |
| TransactionID |
int |
Unique ID for the transaction (= primary key) |
| ParentTransactionID |
int |
The ID of a preceeding transaction of a superior transaction, if transactions are organized in a hierarchy |
| TransactionType |
nvarchar (50) |
Type of the transaction, e.g. gift in or out, exchange in or out, purchase in or out |
| TransactionTitle |
nvarchar (200) |
The title of the transaction as e.g. shown in an user interface |
| ReportingCategory |
nvarchar (50) |
A group defined for the transaction, e.g. a taxonomic group as used for exchange balancing |
| AdministratingCollectionID |
int |
ID of the collection which is responsible for the administration of the transaction. |
| MaterialDescription |
nvarchar (MAX) |
Description of the material of this transaction |
| MaterialSource |
nvarchar (500) |
The source of the material within a transaction, e.g. a excavation |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. |
| MaterialCollectors |
nvarchar (MAX) |
The collectors of the material |
| FromCollectionID |
int |
The ID of the collection from which the specimen were transfered, e.g. the donating collection of a gift |
| FromTransactionPartnerName |
nvarchar (255) |
Name of the person or institution from which the specimen were transfered, e.g. the donator of a gift |
| FromTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
| FromTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the source of the specimen, e.g. the donating collection of a gift |
| ToCollectionID |
int |
The ID of the collection to which the specimen were transfered, e.g. the receiver of a gift |
| ToTransactionPartnerName |
nvarchar (255) |
Name of the person or institution to which the specimen were transfered, e.g. the receiver of a gift |
| ToTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
| ToTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the destination of the specimen, e.g. the receiving collection of a gift |
| ToRecipient |
nvarchar (255) |
The recipient receiving the transaction e.g. if not derived from the link to DiversityAgents |
| NumberOfUnits |
int |
The number of units which were (initially) included in the transaction |
| Investigator |
nvarchar (200) |
The investigator for whose study a transacted material was sent |
| TransactionComment |
nvarchar (MAX) |
Comments on the exchanged material addressed to the transaction partner |
| BeginDate |
datetime |
Date when the transaction started |
| AgreedEndDate |
datetime |
End of the transaction period, e.g. if the time for borrowing the specimen is restricted |
| ActualEndDate |
datetime |
Actual end of the transaction after a prolonation when e.g. the date of return for a loan was prolonged by the owner |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction not to be published e.g. on a web page |
| ResponsibleName |
nvarchar (255) |
The person responsible for this transaction |
| ResponsibleAgentURI |
varchar (255) |
The URI of the person, team or organisation responsible for the data (see e.g. module DiversityAgents) |
| Accessible |
tinyint |
If a user has access to this dataset |
Depending on:
- Transaction
- TransactionChildNodesAccess
Function TransactionCurrency
The default curreny for payments
DataType: nvarchar (50)
Function TransactionHierarchy
Returns a table that lists all the transactions related to the given transaction.
| Parameter |
DataType |
Description |
| @TransactionID |
int |
Unique ID for the transaction (= primary key) |
| Column |
DataType |
Description |
| TransactionID |
int |
Unique ID for the transaction (= primary key) |
| ParentTransactionID |
int |
The ID of a preceeding transaction of a superior transaction, if transactions are organized in a hierarchy |
| TransactionType |
nvarchar (50) |
Type of the transaction, e.g. gift in or out, exchange in or out, purchase in or out |
| TransactionTitle |
nvarchar (200) |
The title of the transaction as e.g. shown in an user interface |
| ReportingCategory |
nvarchar (50) |
A group defined for the transaction, e.g. a taxonomic group as used for exchange balancing |
| AdministratingCollectionID |
int |
ID of the collection which is responsible for the administration of the transaction. |
| MaterialDescription |
nvarchar (MAX) |
Description of the material of this transaction |
| MaterialSource |
nvarchar (500) |
The source of the material within a transaction, e.g. a excavation |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. |
| MaterialCollectors |
nvarchar (MAX) |
The collectors of the material |
| FromCollectionID |
int |
The ID of the collection from which the specimen were transfered, e.g. the donating collection of a gift |
| FromTransactionPartnerName |
nvarchar (255) |
Name of the person or institution from which the specimen were transfered, e.g. the donator of a gift |
| FromTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
| FromTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the source of the specimen, e.g. the donating collection of a gift |
| ToCollectionID |
int |
The ID of the collection to which the specimen were transfered, e.g. the receiver of a gift |
| ToTransactionPartnerName |
nvarchar (255) |
Name of the person or institution to which the specimen were transfered, e.g. the receiver of a gift |
| ToTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
| ToTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the destination of the specimen, e.g. the receiving collection of a gift |
| ToRecipient |
nvarchar (255) |
The recipient receiving the transaction e.g. if not derived from the link to DiversityAgents |
| NumberOfUnits |
int |
The number of units which were (initially) included in the transaction |
| Investigator |
nvarchar (200) |
The investigator for whose study a transacted material was sent |
| TransactionComment |
nvarchar (MAX) |
Comments on the exchanged material addressed to the transaction partner |
| BeginDate |
datetime |
Date when the transaction started |
| AgreedEndDate |
datetime |
End of the transaction period, e.g. if the time for borrowing the specimen is restricted |
| ActualEndDate |
datetime |
Actual end of the transaction after a prolonation when e.g. the date of return for a loan was prolonged by the owner |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction not to be published e.g. on a web page |
| ResponsibleName |
nvarchar (255) |
The person responsible for this transaction |
| ResponsibleAgentURI |
varchar (255) |
The URI of the person, team or organisation responsible for the data (see e.g. module DiversityAgents) |
| DateSupplement |
nvarchar (100) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. |
Depending on:
- TransactionChildNodes
- TransactionList
Function TransactionHierarchyAccess
Returns a table that lists all the transactions related to the given transaction including the accessiblity.
| Parameter |
DataType |
Description |
| @TransactionID |
int |
Unique ID for the transaction (= primary key) |
| Column |
DataType |
Description |
| TransactionID |
int |
Unique ID for the transaction (= primary key) |
| ParentTransactionID |
int |
The ID of a preceeding transaction of a superior transaction, if transactions are organized in a hierarchy |
| TransactionType |
nvarchar (50) |
Type of the transaction, e.g. gift in or out, exchange in or out, purchase in or out |
| TransactionTitle |
nvarchar (200) |
The title of the transaction as e.g. shown in an user interface |
| ReportingCategory |
nvarchar (50) |
A group defined for the transaction, e.g. a taxonomic group as used for exchange balancing |
| AdministratingCollectionID |
int |
ID of the collection which is responsible for the administration of the transaction. |
| MaterialDescription |
nvarchar (MAX) |
Description of the material of this transaction |
| MaterialSource |
nvarchar (500) |
The source of the material within a transaction, e.g. a excavation |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. |
| MaterialCollectors |
nvarchar (MAX) |
The collectors of the material |
| FromCollectionID |
int |
The ID of the collection from which the specimen were transfered, e.g. the donating collection of a gift |
| FromTransactionPartnerName |
nvarchar (255) |
Name of the person or institution from which the specimen were transfered, e.g. the donator of a gift |
| FromTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
| FromTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the source of the specimen, e.g. the donating collection of a gift |
| ToCollectionID |
int |
The ID of the collection to which the specimen were transfered, e.g. the receiver of a gift |
| ToTransactionPartnerName |
nvarchar (255) |
Name of the person or institution to which the specimen were transfered, e.g. the receiver of a gift |
| ToTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
| ToTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the destination of the specimen, e.g. the receiving collection of a gift |
| ToRecipient |
nvarchar (255) |
The recipient receiving the transaction e.g. if not derived from the link to DiversityAgents |
| NumberOfUnits |
int |
The number of units which were (initially) included in the transaction |
| Investigator |
nvarchar (200) |
The investigator for whose study a transacted material was sent |
| TransactionComment |
nvarchar (MAX) |
Comments on the exchanged material addressed to the transaction partner |
| BeginDate |
datetime |
Date when the transaction started |
| AgreedEndDate |
datetime |
End of the transaction period, e.g. if the time for borrowing the specimen is restricted |
| ActualEndDate |
datetime |
Actual end of the transaction after a prolonation when e.g. the date of return for a loan was prolonged by the owner |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction not to be published e.g. on a web page |
| ResponsibleName |
nvarchar (255) |
The person responsible for this transaction |
| ResponsibleAgentURI |
varchar (255) |
The URI of the person, team or organisation responsible for the data (see e.g. module DiversityAgents) |
| Accessible |
tinyint |
If a user has access to this dataset |
Depending on:
- Transaction
- TransactionChildNodesAccess
- TransactionList
Function TransactionHierarchyAll
Returns a table that lists all the transactions including their hierarchy
| Column |
DataType |
Description |
| TransactionID |
int |
Unique ID for the transaction (= primary key) |
| ParentTransactionID |
int |
The ID of a preceeding transaction of a superior transaction, if transactions are organized in a hierarchy |
| TransactionType |
nvarchar (50) |
Type of the transaction, e.g. gift in or out, exchange in or out, purchase in or out |
| TransactionTitle |
nvarchar (200) |
The title of the transaction as e.g. shown in an user interface |
| ReportingCategory |
nvarchar (50) |
A group defined for the transaction, e.g. a taxonomic group as used for exchange balancing |
| AdministratingCollectionID |
int |
ID of the collection which is responsible for the administration of the transaction. |
| MaterialDescription |
nvarchar (MAX) |
Description of the material of this transaction |
| MaterialSource |
nvarchar (500) |
The source of the material within a transaction, e.g. a excavation |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. |
| MaterialCollectors |
nvarchar (MAX) |
The collectors of the material |
| FromCollectionID |
int |
The ID of the collection from which the specimen were transfered, e.g. the donating collection of a gift |
| FromTransactionPartnerName |
nvarchar (255) |
Name of the person or institution from which the specimen were transfered, e.g. the donator of a gift |
| FromTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
| FromTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the source of the specimen, e.g. the donating collection of a gift |
| ToCollectionID |
int |
The ID of the collection to which the specimen were transfered, e.g. the receiver of a gift |
| ToTransactionPartnerName |
nvarchar (255) |
Name of the person or institution to which the specimen were transfered, e.g. the receiver of a gift |
| ToTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
| ToTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the destination of the specimen, e.g. the receiving collection of a gift |
| ToRecipient |
nvarchar (255) |
The recipient receiving the transaction e.g. if not derived from the link to DiversityAgents |
| NumberOfUnits |
smallint |
The number of units which were (initially) included in the transaction |
| Investigator |
nvarchar (200) |
The investigator for whose study a transacted material was sent |
| TransactionComment |
nvarchar (MAX) |
Comments on the exchanged material addressed to the transaction partner |
| BeginDate |
datetime |
Date when the transaction started |
| AgreedEndDate |
datetime |
End of the transaction period, e.g. if the time for borrowing the specimen is restricted |
| ActualEndDate |
datetime |
Actual end of the transaction after a prolonation when e.g. the date of return for a loan was prolonged by the owner |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction not to be published e.g. on a web page |
| ResponsibleName |
nvarchar (255) |
The person responsible for this transaction |
| ResponsibleAgentURI |
varchar (255) |
The URI of the person, team or organisation responsible for the data (see e.g. module DiversityAgents) |
| DisplayText |
nvarchar (500) |
Hierarchy of the transaction starting at top transaction separated by |
| HierarchyDisplayText |
nvarchar (500) |
Hierarchy of the transaction starting at top transaction separated by |
Depending on:
Function UserCollectionList
Returns a table that lists all the collections a User has access to, including the child collections.
| Column |
DataType |
Description |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
| CollectionName |
nvarchar (255) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
| CollectionAcronym |
nvarchar (50) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
| AdministrativeContactAgentURI |
nvarchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
| Description |
nvarchar (MAX) |
A short description of the collection |
| Location |
nvarchar (1000) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
Depending on:
- Collection
- CollectionChildNodes
- CollectionManager
- CollectionUser
- ManagerCollectionList
Function UserID
ID of the User as stored in table UserProxy
DataType: int
Depending on:
Function Version
Version of the database
DataType: nvarchar (8)
Function VersionClient
Minimal version of the client compatible with the database
DataType: nvarchar (11)
PROCEDURES
Procedure DeleteXmlAttribute
Deleting an attribute of an XML node in a column of datatype XML
| Parameter |
DataType |
Description |
| @Table |
nvarchar (128) |
Name of the table containing the XML column |
| @Column |
nvarchar (128) |
Name of the XML column |
| @Path |
nvarchar (4000) |
Path of the XML node |
| @Attribute |
nvarchar (128) |
Attribute that should be removed |
| @WhereClause |
nvarchar (4000) |
Where clause to select the data within the table |
Procedure DeleteXmlNode
Deleting a XML node in a column of datatype XML
| Parameter |
DataType |
Description |
| @Table |
nvarchar (128) |
Name of the table containing the XML column |
| @Column |
nvarchar (128) |
Name of the XML column |
| @Path |
nvarchar (4000) |
Path of the XML node |
| @WhereClause |
nvarchar (4000) |
Where clause to select the data within the table |
Procedure procCopyCollectionSpecimen2
Copy a collection specimen
| Parameter |
DataType |
Description |
| @CollectionSpecimenID |
int |
Refers to ID of CollectionEvent (= foreign key and part of primary key) |
| @OriginalCollectionSpecimenID |
int |
The CollectionSpecimenID of the CollectionSpecimen that should be copied |
| @AccessionNumber |
nvarchar (50) |
Accession number of the specimen within the collection, e.g. “M-29834752” |
| @EventCopyMode |
int |
3 Options: -1 = dont copy the event, leave the entry in table CollectionSpecimen empty; 0 = take same event as original specimen; 1 = create new event with the same data as the old specimen |
| @IncludedTables |
nvarchar (4000) |
Contains list of tables that are copied according to the users choice |
Depending on:
- Annotation
- CollectionAgent
- CollectionEvent
- CollectionEventImage
- CollectionEventLocalisation
- CollectionEventMethod
- CollectionEventParameterValue
- CollectionEventProperty
- CollectionProject
- CollectionSpecimen
- CollectionSpecimenImage
- CollectionSpecimenImageProperty
- CollectionSpecimenPart
- CollectionSpecimenPartDescription
- CollectionSpecimenProcessing
- CollectionSpecimenProcessingMethod
- CollectionSpecimenProcessingMethodParameter
- CollectionSpecimenReference
- CollectionSpecimenRelation
- CollectionSpecimenTransaction
- ExternalIdentifier
- Identification
- IdentificationUnit
- IdentificationUnitAnalysis
- IdentificationUnitAnalysisMethod
- IdentificationUnitAnalysisMethodParameter
- IdentificationUnitGeoAnalysis
- IdentificationUnitInPart
Procedure procCopyCollectionSpecimenPart
Copy a collection specimen part
| Parameter |
DataType |
Description |
| @SpecimenPartID |
int |
Unique ID of the part of the collection specimen (= part of primary key). |
| @OriginalSpecimenPartID |
int |
The SpecimenPartID of the CollectionSpecimenPart that should be copied |
| @AccessionNumber |
nvarchar (50) |
Accession number of the part of the specimen within the collection, if it is different from the accession number of the specimen as stored in the table CollectionSpecimen, e.g. “M-29834752” |
| @PartSublabel |
nvarchar (50) |
The label for a part of a specimen, e.g. “cone”, or a number attached to a duplicate of a specimen |
| @StorageLocation |
nvarchar (255) |
A code identifying the place where the specimen is stored within the collection. Frequently the accepted scientific name is used as storage location code. |
| @IncludedTables |
nvarchar (4000) |
Contains list of tables that are copied according to the users choice |
Depending on:
- Annotation
- CollectionSpecimenPart
- CollectionSpecimenPartDescription
- CollectionSpecimenPartRegulation
- CollectionSpecimenProcessing
- CollectionSpecimenProcessingMethod
- CollectionSpecimenProcessingMethodParameter
- CollectionSpecimenReference
- CollectionSpecimenRelation
- CollectionSpecimenTransaction
- ExternalIdentifier
- IdentificationUnitInPart
Procedure procFillCacheDescription
Filling table CacheDescription
Depending on:
- CacheDescription
- EntityRepresentation
Procedure procInsertCollectionEventCopy
Copy a collection event
| Parameter |
DataType |
Description |
| @CollectionEventID |
int |
Unique ID for the table CollectionEvent (= primary key) |
| @OriginalCollectionEventID |
int |
The CollectionEventID of the CollectionEvent that should be copied |
Depending on:
- CollectionEvent
- CollectionEventImage
- CollectionEventLocalisation
- CollectionEventProperty
Procedure procSetVersionCollectionEvent
Setting the version of a dataset
| Parameter |
DataType |
Description |
| @ID |
int |
CollectionEventID of the dataset |
Depending on:
- CollectionEvent
- CollectionProject
- CollectionSpecimen
- ProjectProxy
Procedure procSetVersionCollectionSpecimen
Setting the version of a dataset
| Parameter |
DataType |
Description |
| @ID |
int |
CollectionSpecimenID of the dataset |
Depending on:
- CollectionProject
- CollectionSpecimen
- ProjectProxy
Procedure SetXmlAttribute
Setting a value of an XML node
| Parameter |
DataType |
Description |
| @Table |
nvarchar (128) |
Name of the table containing the XML column |
| @Column |
nvarchar (128) |
Name of the XML column |
| @Path |
nvarchar (4000) |
Path of the XML node |
| @Attribute |
nvarchar (128) |
Attribute that should be set |
| @Value |
nvarchar (4000) |
The value for the attribute that should be set |
| @WhereClause |
nvarchar (4000) |
Where clause to select the data within the table |
Procedure SetXmlValue
Setting a value of an XML node
| Parameter |
DataType |
Description |
| @Table |
nvarchar (128) |
Name of the table containing the XML column |
| @Column |
nvarchar (128) |
Name of the XML column |
| @Path |
nvarchar (4000) |
Path of the XML node |
| @Value |
nvarchar (4000) |
The value for the node that should be set |
| @WhereClause |
nvarchar (4000) |
Where clause to select the data within the table |
Diversity Collection
Roles
| Content of cell |
Permission |
|
Not granted |
| Name of other role |
Inherited from other role |
| • |
Granted |
Role Administrator
Administrator of the database. Read/write access to all objects
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
Editor |
Data Manager |
Data Manager |
• |
|
TABLE |
| Analysis_log |
|
Data Manager |
|
|
|
TABLE |
| AnalysisResult |
User |
• |
• |
• |
|
TABLE |
| AnalysisTaxonomicGroup |
User |
• |
• |
• |
|
TABLE |
| Annotation |
User |
User |
Data Manager |
Data Manager |
|
TABLE |
| Annotation_log |
Editor |
Editor |
|
|
|
TABLE |
| AnnotationType_Enum |
User |
|
|
|
|
TABLE |
| AnonymCollector |
Cache User |
Cache Admin |
Cache Admin |
Cache Admin |
|
TABLE |
| CacheDatabase2 |
Cache User |
Cache Admin |
|
|
|
TABLE |
| CacheDescription |
User |
User |
User |
User |
|
TABLE |
| CollCircumstances_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
User |
• |
• |
• |
|
TABLE |
| CollCollectionType_Enum |
User |
• |
• |
• |
|
TABLE |
| CollDateCategory_Enum |
User |
• |
• |
• |
|
TABLE |
| Collection |
Editor |
• |
Editor |
• |
|
TABLE |
| Collection_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionAgent |
User |
Editor |
Editor |
• |
|
TABLE |
| CollectionAgent_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionEvent |
User |
Editor |
Editor |
• |
|
TABLE |
| CollectionEvent_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionEventImage |
User |
Editor |
Editor |
• |
|
TABLE |
| CollectionEventImage_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionEventLocalisation |
User |
Typist |
Typist |
Editor |
|
TABLE |
| CollectionEventLocalisation_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionEventMethod |
User |
Data Manager |
Data Manager |
Data Manager |
|
TABLE |
| CollectionEventMethod_log |
|
• |
|
|
|
TABLE |
| CollectionEventParameterValue |
User |
Data Manager |
Data Manager |
Data Manager |
|
TABLE |
| CollectionEventParameterValue_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionEventProperty |
User |
Typist |
Typist |
Editor |
|
TABLE |
| CollectionEventProperty_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionEventRegulation |
User |
Editor |
Editor |
• |
|
TABLE |
| CollectionEventRegulation_log |
Editor |
Editor |
|
• |
|
TABLE |
| CollectionEventSeries |
User |
Editor |
Typist |
Editor |
|
TABLE |
| CollectionEventSeries_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
User |
Editor |
Typist |
Editor |
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
User |
• |
• |
• |
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
• |
• |
• |
• |
|
TABLE |
| CollectionImage_log |
• |
• |
|
|
|
TABLE |
| CollectionManager |
Collection Manager |
• |
• |
• |
|
TABLE |
| CollectionProject |
User |
Editor |
Editor |
• |
|
TABLE |
| CollectionProject_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimen |
Typist |
Editor |
Editor |
• |
|
TABLE |
| CollectionSpecimen_log |
Editor |
Editor |
|
• |
|
TABLE |
| CollectionSpecimenImage |
Typist |
Editor |
Editor |
• |
|
TABLE |
| CollectionSpecimenImage_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenImageProperty_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimenPart |
User |
Typist |
Typist |
Editor |
|
TABLE |
| CollectionSpecimenPart_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenPartDescription_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimenProcessing |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenProcessing_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
Editor |
Editor |
|
• |
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenReference_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimenRelation |
User |
Editor |
Editor |
• |
|
TABLE |
| CollectionSpecimenRelation_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimenTransaction |
User |
Typist |
Typist |
Editor |
|
TABLE |
| CollectionSpecimenTransaction_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionTask |
User |
Collection Manager |
Collection Manager |
Collection Manager |
|
TABLE |
| CollectionTask_log |
Collection Manager |
Collection Manager |
|
|
|
TABLE |
| CollectionTaskImage |
User |
Collection Manager |
Collection Manager |
Collection Manager |
|
TABLE |
| CollectionTaskImage_log |
Collection Manager |
Collection Manager |
|
|
|
TABLE |
| CollectionTaskMetric |
User |
Collection Manager |
Collection Manager |
Collection Manager |
|
TABLE |
| CollectionTaskMetric_log |
Collection Manager |
Collection Manager |
|
|
|
TABLE |
| CollEventDateCategory_Enum |
Editor |
• |
• |
• |
|
TABLE |
| CollEventImageType_Enum |
Editor |
• |
• |
• |
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
User |
|
|
|
|
TABLE |
| CollExchangeType_Enum |
Editor |
• |
• |
• |
|
TABLE |
| CollIdentificationCategory_Enum |
Editor |
• |
• |
• |
|
TABLE |
| CollIdentificationDateCategory_Enum |
User |
• |
• |
• |
|
TABLE |
| CollIdentificationQualifier_Enum |
User |
• |
• |
• |
|
TABLE |
| CollLabelTranscriptionState_Enum |
User |
• |
• |
• |
|
TABLE |
| CollLabelType_Enum |
Editor |
• |
• |
• |
|
TABLE |
| CollMaterialCategory_Enum |
User |
• |
• |
• |
|
TABLE |
| CollRetrievalType_Enum |
User |
• |
• |
• |
|
TABLE |
| CollSpecimenImageType_Enum |
User |
• |
• |
• |
|
TABLE |
| CollSpecimenRelationType_Enum |
User |
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
User |
• |
• |
• |
|
TABLE |
| CollTaxonomicGroup_Enum |
User |
• |
• |
• |
|
TABLE |
| CollTransactionType_Enum |
User |
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
User |
• |
• |
• |
|
TABLE |
| CollUnitRelationType_Enum |
User |
|
|
|
|
TABLE |
| Entity |
User |
• |
• |
• |
|
TABLE |
| EntityAccessibility_Enum |
• |
|
|
|
|
TABLE |
| EntityContext_Enum |
User |
• |
• |
• |
|
TABLE |
| EntityDetermination_Enum |
User |
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
User |
• |
• |
• |
|
TABLE |
| EntityRepresentation |
User |
• |
• |
• |
|
TABLE |
| EntityUsage |
User |
• |
• |
• |
|
TABLE |
| EntityUsage_Enum |
User |
• |
• |
• |
|
TABLE |
| EntityVisibility_Enum |
User |
|
|
|
|
TABLE |
| ExternalIdentifier |
User |
Editor |
Editor |
Editor |
|
TABLE |
| ExternalIdentifier_log |
Editor |
Editor |
|
|
|
TABLE |
| ExternalIdentifierType |
User |
• |
• |
• |
|
TABLE |
| ExternalIdentifierType_log |
• |
• |
|
|
|
TABLE |
| Identification |
User |
Typist |
Typist |
• |
|
TABLE |
| Identification_log |
Editor |
Editor |
|
|
|
TABLE |
| IdentificationUnit |
User |
Editor |
Editor |
• |
|
TABLE |
| IdentificationUnit_log |
Editor |
Editor |
|
|
|
TABLE |
| IdentificationUnitAnalysis |
User |
Editor |
Editor |
• |
|
TABLE |
| IdentificationUnitAnalysis_log |
Typist |
Typist |
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
User |
Editor |
Editor |
Editor |
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
Editor |
Editor |
|
• |
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
User |
Editor |
Editor |
Editor |
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
Editor |
Editor |
|
• |
|
TABLE |
| IdentificationUnitGeoAnalysis |
User |
Editor |
Editor |
Editor |
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
Editor |
|
|
|
TABLE |
| IdentificationUnitInPart |
User |
Typist |
Typist |
Editor |
|
TABLE |
| IdentificationUnitInPart_log |
Typist |
Typist |
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
User |
• |
• |
• |
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
User |
• |
• |
• |
|
TABLE |
| Method_log |
|
• |
|
|
|
TABLE |
| MethodForAnalysis |
User |
• |
• |
• |
|
TABLE |
| MethodForProcessing |
User |
• |
• |
• |
|
TABLE |
| Parameter |
User |
• |
• |
• |
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
User |
• |
• |
• |
|
TABLE |
| Processing |
User |
Data Manager |
Data Manager |
• |
|
TABLE |
| Processing_log |
Editor |
Data Manager |
|
|
|
TABLE |
| ProcessingMaterialCategory |
User |
Data Manager |
Data Manager |
• |
|
TABLE |
| ProjectAnalysis |
User |
Data Manager |
Data Manager |
• |
|
TABLE |
| ProjectMaterialCategory |
User |
• |
• |
• |
|
TABLE |
| ProjectProcessing |
User |
Data Manager |
Data Manager |
• |
|
TABLE |
| ProjectTaxonomicGroup |
User |
• |
• |
• |
|
TABLE |
| ProjectUser |
• |
• |
• |
• |
|
TABLE |
| Property |
User |
• |
• |
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
User |
• |
• |
|
|
TABLE |
| ReplicationPublisher |
Editor |
• |
|
• |
|
TABLE |
| Task |
User |
Collection Manager |
Collection Manager |
• |
|
TABLE |
| Task_log |
Collection Manager |
Collection Manager |
|
|
|
TABLE |
| TaskDateType_Enum |
User |
|
|
|
|
TABLE |
| TaskModule |
User |
Collection Manager |
Collection Manager |
Collection Manager |
|
TABLE |
| TaskModule_log |
Collection Manager |
Collection Manager |
|
|
|
TABLE |
| TaskModuleType_Enum |
User |
|
|
|
|
TABLE |
| TaskResult |
User |
Collection Manager |
Collection Manager |
Collection Manager |
|
TABLE |
| TaskResult_log |
Collection Manager |
Collection Manager |
|
|
|
TABLE |
| TaskType_Enum |
User |
• |
• |
• |
|
TABLE |
| Transaction |
User |
Collection Manager |
Collection Manager |
• |
|
TABLE |
| Transaction_log |
Typist |
Typist |
|
|
|
TABLE |
| TransactionAgent |
Transaction User |
Collection Manager |
Collection Manager |
Collection Manager |
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
User |
• |
• |
• |
|
TABLE |
| TransactionDocument |
User |
Typist |
Typist |
• |
|
TABLE |
| TransactionDocument_log |
Typist |
Typist |
|
|
|
TABLE |
| TransactionPayment |
Transaction User |
Collection Manager |
Collection Manager |
Collection Manager |
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
User |
|
|
|
|
VIEW |
| AnnotationPart |
User |
|
|
|
|
VIEW |
| AnnotationSpecimen |
User |
|
|
|
|
VIEW |
| AnnotationUnit |
User |
|
|
|
|
VIEW |
| CollectionAgent_Core |
User |
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
User |
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
User |
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
User |
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
|
|
|
|
|
VIEW |
| FirstLinesIdentification |
User |
|
|
|
|
VIEW |
| Identification_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
User |
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
User |
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
User |
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
User |
|
|
|
|
VIEW |
| TransactionList_H7 |
User |
|
|
|
|
VIEW |
| TransactionPermit |
User |
|
|
|
|
VIEW |
| TransactionRegulation |
User |
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
User |
|
|
|
|
VIEW |
| ViewCollectionEventImage |
User |
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
User |
|
|
|
|
VIEW |
| ViewCollectionImage |
User |
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
User |
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
User |
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
User |
|
|
|
|
VIEW |
| AgentOneString |
Editor |
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
User |
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
User |
|
|
|
|
FUNCTION |
| AnalysisList |
User |
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
User |
|
|
|
|
FUNCTION |
| AnalysisProjectList |
User |
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
User |
FUNCTION |
| CollCharacterType_List |
Editor |
|
|
|
|
FUNCTION |
| CollDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollectionChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
User |
|
|
|
|
FUNCTION |
| CollectionLocation |
User |
|
|
|
|
FUNCTION |
| CollectionLocationAll |
User |
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
User |
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
User |
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
User |
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
Editor |
|
|
|
|
FUNCTION |
| CollEventImageType_List |
Editor |
|
|
|
|
FUNCTION |
| CollExchangeType_List |
Editor |
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
Editor |
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
User |
|
|
|
|
FUNCTION |
| CollLabelType_List |
User |
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
User |
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
User |
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
User |
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
User |
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
Editor |
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
Collection Manager |
|
|
|
|
FUNCTION |
| CurrentUser |
User |
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
User |
FUNCTION |
| DefaultProjectID |
|
|
|
|
User |
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
User |
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
User |
FUNCTION |
| EntityInformation_2 |
User |
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
User |
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
User |
FUNCTION |
| EventSeriesChildNodes |
User |
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
User |
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
User |
FUNCTION |
| EventSuperiorList |
User |
|
|
|
|
FUNCTION |
| FirstLines_4 |
User |
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesSeries |
User |
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
User |
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
User |
|
|
|
|
FUNCTION |
| ManagerCollectionList |
Collection Manager |
|
|
|
|
FUNCTION |
| MethodChildNodes |
User |
|
|
|
|
FUNCTION |
| MethodHierarchy |
User |
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
User |
|
|
|
|
FUNCTION |
| NameList |
User |
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
User |
|
|
|
|
FUNCTION |
| NameListPlants |
User |
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
Typist |
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
Editor |
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
User |
FUNCTION |
| ProcessingChildNodes |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
User |
|
|
|
|
FUNCTION |
| ProcessingListForPart |
User |
|
|
|
|
FUNCTION |
| ProcessingProjectList |
User |
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
User |
FUNCTION |
| RequesterCollectionList |
|
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
User |
FUNCTION |
| TaskChildNodes |
User |
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaskHierarchy |
User |
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
User |
FUNCTION |
| TransactionChildNodes |
User |
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
User |
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
User |
FUNCTION |
| TransactionHierarchy |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| UserCollectionList |
User |
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
User |
FUNCTION |
| Version |
|
|
|
|
User |
FUNCTION |
| VersionClient |
|
|
|
|
User |
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
User |
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
Editor |
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
Editor |
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
User |
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
Editor |
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
Editor |
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| SetXmlValue |
|
|
|
|
User |
PROCEDURE |
| Inheriting from roles: |
|
|
|
|
|
|
- Replicator
- DataManager
- CacheAdmin
- CollectionManager
- Editor
Role AdminNonProject
Permissions as Administrator without permission to change projects
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
Editor |
Data Manager |
Data Manager |
Administrator |
|
TABLE |
| Analysis_log |
|
Data Manager |
|
|
|
TABLE |
| AnalysisResult |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| AnalysisTaxonomicGroup |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| Annotation |
User |
User |
Data Manager |
Data Manager |
|
TABLE |
| Annotation_log |
Editor |
Editor |
|
|
|
TABLE |
| AnnotationType_Enum |
User |
|
|
|
|
TABLE |
| AnonymCollector |
Cache User |
Cache Admin |
Cache Admin |
Cache Admin |
|
TABLE |
| CacheDatabase2 |
Cache User |
Cache Admin |
|
|
|
TABLE |
| CacheDescription |
User |
User |
User |
User |
|
TABLE |
| CollCircumstances_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollCollectionType_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollDateCategory_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| Collection |
Editor |
Administrator |
Editor |
Administrator |
|
TABLE |
| Collection_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionAgent |
User |
Editor |
Editor |
Administrator |
|
TABLE |
| CollectionAgent_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionEvent |
User |
Editor |
Editor |
Administrator |
|
TABLE |
| CollectionEvent_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionEventImage |
User |
Editor |
Editor |
Administrator |
|
TABLE |
| CollectionEventImage_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionEventLocalisation |
User |
Typist |
Typist |
Editor |
|
TABLE |
| CollectionEventLocalisation_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionEventMethod |
User |
Data Manager |
Data Manager |
Data Manager |
|
TABLE |
| CollectionEventMethod_log |
|
Administrator |
|
|
|
TABLE |
| CollectionEventParameterValue |
User |
Data Manager |
Data Manager |
Data Manager |
|
TABLE |
| CollectionEventParameterValue_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionEventProperty |
User |
Typist |
Typist |
Editor |
|
TABLE |
| CollectionEventProperty_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionEventRegulation |
User |
Editor |
Editor |
Administrator |
|
TABLE |
| CollectionEventRegulation_log |
Editor |
Editor |
|
Administrator |
|
TABLE |
| CollectionEventSeries |
User |
Editor |
Typist |
Editor |
|
TABLE |
| CollectionEventSeries_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
User |
Editor |
Typist |
Editor |
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
Administrator |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollectionImage_log |
Administrator |
Administrator |
|
|
|
TABLE |
| CollectionManager |
Collection Manager |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollectionProject |
User |
Editor |
Editor |
Administrator |
|
TABLE |
| CollectionProject_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimen |
Typist |
Editor |
Editor |
Administrator |
|
TABLE |
| CollectionSpecimen_log |
Editor |
Editor |
|
Administrator |
|
TABLE |
| CollectionSpecimenImage |
Typist |
Editor |
Editor |
Administrator |
|
TABLE |
| CollectionSpecimenImage_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenImageProperty_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimenPart |
User |
Typist |
Typist |
Editor |
|
TABLE |
| CollectionSpecimenPart_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenPartDescription_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimenProcessing |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenProcessing_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
Editor |
Editor |
|
Administrator |
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenReference_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimenRelation |
User |
Editor |
Editor |
Administrator |
|
TABLE |
| CollectionSpecimenRelation_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimenTransaction |
User |
Typist |
Typist |
Editor |
|
TABLE |
| CollectionSpecimenTransaction_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionTask |
User |
Collection Manager |
Collection Manager |
Collection Manager |
|
TABLE |
| CollectionTask_log |
Collection Manager |
Collection Manager |
|
|
|
TABLE |
| CollectionTaskImage |
User |
Collection Manager |
Collection Manager |
Collection Manager |
|
TABLE |
| CollectionTaskImage_log |
Collection Manager |
Collection Manager |
|
|
|
TABLE |
| CollectionTaskMetric |
User |
Collection Manager |
Collection Manager |
Collection Manager |
|
TABLE |
| CollectionTaskMetric_log |
Collection Manager |
Collection Manager |
|
|
|
TABLE |
| CollEventDateCategory_Enum |
Editor |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollEventImageType_Enum |
Editor |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
User |
|
|
|
|
TABLE |
| CollExchangeType_Enum |
Editor |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollIdentificationCategory_Enum |
Editor |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollIdentificationDateCategory_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollIdentificationQualifier_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollLabelTranscriptionState_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollLabelType_Enum |
Editor |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollMaterialCategory_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollRetrievalType_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollSpecimenImageType_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollSpecimenRelationType_Enum |
User |
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollTaxonomicGroup_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollTransactionType_Enum |
User |
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| CollUnitRelationType_Enum |
User |
|
|
|
|
TABLE |
| Entity |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| EntityAccessibility_Enum |
Administrator |
|
|
|
|
TABLE |
| EntityContext_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| EntityDetermination_Enum |
User |
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| EntityRepresentation |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| EntityUsage |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| EntityUsage_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| EntityVisibility_Enum |
User |
|
|
|
|
TABLE |
| ExternalIdentifier |
User |
Editor |
Editor |
Editor |
|
TABLE |
| ExternalIdentifier_log |
Editor |
Editor |
|
|
|
TABLE |
| ExternalIdentifierType |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| ExternalIdentifierType_log |
Administrator |
Administrator |
|
|
|
TABLE |
| Identification |
User |
Typist |
Typist |
Administrator |
|
TABLE |
| Identification_log |
Editor |
Editor |
|
|
|
TABLE |
| IdentificationUnit |
User |
Editor |
Editor |
Administrator |
|
TABLE |
| IdentificationUnit_log |
Editor |
Editor |
|
|
|
TABLE |
| IdentificationUnitAnalysis |
User |
Editor |
Editor |
Administrator |
|
TABLE |
| IdentificationUnitAnalysis_log |
Typist |
Typist |
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
User |
Editor |
Editor |
Editor |
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
Editor |
Editor |
|
Administrator |
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
User |
Editor |
Editor |
Editor |
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
Editor |
Editor |
|
Administrator |
|
TABLE |
| IdentificationUnitGeoAnalysis |
User |
Editor |
Editor |
Editor |
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
Editor |
|
|
|
TABLE |
| IdentificationUnitInPart |
User |
Typist |
Typist |
Editor |
|
TABLE |
| IdentificationUnitInPart_log |
Typist |
Typist |
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| Method_log |
|
Administrator |
|
|
|
TABLE |
| MethodForAnalysis |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| MethodForProcessing |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| Parameter |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| Processing |
User |
Data Manager |
Data Manager |
Administrator |
|
TABLE |
| Processing_log |
Editor |
Data Manager |
|
|
|
TABLE |
| ProcessingMaterialCategory |
User |
Data Manager |
Data Manager |
Administrator |
|
TABLE |
| ProjectAnalysis |
User |
Data Manager |
Data Manager |
Administrator |
|
TABLE |
| ProjectMaterialCategory |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| ProjectProcessing |
User |
Data Manager |
Data Manager |
Administrator |
|
TABLE |
| ProjectTaxonomicGroup |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| ProjectUser |
Administrator |
Administrator |
Administrator |
Administrator |
|
TABLE |
| Property |
User |
Administrator |
Administrator |
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
User |
Administrator |
Administrator |
|
|
TABLE |
| ReplicationPublisher |
Editor |
Administrator |
|
Administrator |
|
TABLE |
| Task |
User |
Collection Manager |
Collection Manager |
Administrator |
|
TABLE |
| Task_log |
Collection Manager |
Collection Manager |
|
|
|
TABLE |
| TaskDateType_Enum |
User |
|
|
|
|
TABLE |
| TaskModule |
User |
Collection Manager |
Collection Manager |
Collection Manager |
|
TABLE |
| TaskModule_log |
Collection Manager |
Collection Manager |
|
|
|
TABLE |
| TaskModuleType_Enum |
User |
|
|
|
|
TABLE |
| TaskResult |
User |
Collection Manager |
Collection Manager |
Collection Manager |
|
TABLE |
| TaskResult_log |
Collection Manager |
Collection Manager |
|
|
|
TABLE |
| TaskType_Enum |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| Transaction |
User |
Collection Manager |
Collection Manager |
Administrator |
|
TABLE |
| Transaction_log |
Typist |
Typist |
|
|
|
TABLE |
| TransactionAgent |
Transaction User |
Collection Manager |
Collection Manager |
Collection Manager |
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
User |
Administrator |
Administrator |
Administrator |
|
TABLE |
| TransactionDocument |
User |
Typist |
Typist |
Administrator |
|
TABLE |
| TransactionDocument_log |
Typist |
Typist |
|
|
|
TABLE |
| TransactionPayment |
Transaction User |
Collection Manager |
Collection Manager |
Collection Manager |
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
User |
|
|
|
|
VIEW |
| AnnotationPart |
User |
|
|
|
|
VIEW |
| AnnotationSpecimen |
User |
|
|
|
|
VIEW |
| AnnotationUnit |
User |
|
|
|
|
VIEW |
| CollectionAgent_Core |
User |
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
User |
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
User |
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
User |
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
|
|
|
|
|
VIEW |
| FirstLinesIdentification |
User |
|
|
|
|
VIEW |
| Identification_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
User |
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
User |
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
User |
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
User |
|
|
|
|
VIEW |
| TransactionList_H7 |
User |
|
|
|
|
VIEW |
| TransactionPermit |
User |
|
|
|
|
VIEW |
| TransactionRegulation |
User |
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
User |
|
|
|
|
VIEW |
| ViewCollectionEventImage |
User |
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
User |
|
|
|
|
VIEW |
| ViewCollectionImage |
User |
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
User |
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
User |
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
User |
|
|
|
|
VIEW |
| AgentOneString |
Editor |
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
User |
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
User |
|
|
|
|
FUNCTION |
| AnalysisList |
User |
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
User |
|
|
|
|
FUNCTION |
| AnalysisProjectList |
User |
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
User |
FUNCTION |
| CollCharacterType_List |
Editor |
|
|
|
|
FUNCTION |
| CollDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollectionChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
User |
|
|
|
|
FUNCTION |
| CollectionLocation |
User |
|
|
|
|
FUNCTION |
| CollectionLocationAll |
User |
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
User |
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
User |
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
User |
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
Editor |
|
|
|
|
FUNCTION |
| CollEventImageType_List |
Editor |
|
|
|
|
FUNCTION |
| CollExchangeType_List |
Editor |
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
Editor |
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
User |
|
|
|
|
FUNCTION |
| CollLabelType_List |
User |
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
User |
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
User |
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
User |
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
User |
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
Editor |
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
Collection Manager |
|
|
|
|
FUNCTION |
| CurrentUser |
User |
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
User |
FUNCTION |
| DefaultProjectID |
|
|
|
|
User |
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
User |
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
User |
FUNCTION |
| EntityInformation_2 |
User |
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
User |
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
User |
FUNCTION |
| EventSeriesChildNodes |
User |
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
User |
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
User |
FUNCTION |
| EventSuperiorList |
User |
|
|
|
|
FUNCTION |
| FirstLines_4 |
User |
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesSeries |
User |
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
User |
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
User |
|
|
|
|
FUNCTION |
| ManagerCollectionList |
Collection Manager |
|
|
|
|
FUNCTION |
| MethodChildNodes |
User |
|
|
|
|
FUNCTION |
| MethodHierarchy |
User |
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
User |
|
|
|
|
FUNCTION |
| NameList |
User |
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
User |
|
|
|
|
FUNCTION |
| NameListPlants |
User |
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
Typist |
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
Editor |
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
User |
FUNCTION |
| ProcessingChildNodes |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
User |
|
|
|
|
FUNCTION |
| ProcessingListForPart |
User |
|
|
|
|
FUNCTION |
| ProcessingProjectList |
User |
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
User |
FUNCTION |
| RequesterCollectionList |
|
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
User |
FUNCTION |
| TaskChildNodes |
User |
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaskHierarchy |
User |
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
User |
FUNCTION |
| TransactionChildNodes |
User |
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
User |
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
User |
FUNCTION |
| TransactionHierarchy |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| UserCollectionList |
User |
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
User |
FUNCTION |
| Version |
|
|
|
|
User |
FUNCTION |
| VersionClient |
|
|
|
|
User |
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
User |
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
Editor |
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
Editor |
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
User |
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
Editor |
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
Editor |
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| SetXmlValue |
|
|
|
|
User |
PROCEDURE |
| Inheriting from roles: |
|
|
|
|
|
|
Role CacheAdmin
Read/write access to objects related to cache database
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
User |
|
|
|
|
TABLE |
| Analysis_log |
|
|
|
|
|
TABLE |
| AnalysisResult |
User |
|
|
|
|
TABLE |
| AnalysisTaxonomicGroup |
User |
|
|
|
|
TABLE |
| Annotation |
User |
User |
User |
|
|
TABLE |
| Annotation_log |
|
|
|
|
|
TABLE |
| AnnotationType_Enum |
User |
|
|
|
|
TABLE |
| AnonymCollector |
Cache User |
• |
• |
• |
|
TABLE |
| CacheDatabase2 |
Cache User |
• |
|
|
|
TABLE |
| CacheDescription |
User |
User |
User |
User |
|
TABLE |
| CollCircumstances_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionType_Enum |
User |
|
|
|
|
TABLE |
| CollDateCategory_Enum |
User |
|
|
|
|
TABLE |
| Collection |
User |
|
|
|
|
TABLE |
| Collection_log |
|
|
|
|
|
TABLE |
| CollectionAgent |
User |
|
|
|
|
TABLE |
| CollectionAgent_log |
|
|
|
|
|
TABLE |
| CollectionEvent |
User |
|
|
|
|
TABLE |
| CollectionEvent_log |
|
|
|
|
|
TABLE |
| CollectionEventImage |
User |
|
|
|
|
TABLE |
| CollectionEventImage_log |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation |
User |
|
|
|
|
TABLE |
| CollectionEventLocalisation_log |
|
|
|
|
|
TABLE |
| CollectionEventMethod |
User |
|
|
|
|
TABLE |
| CollectionEventMethod_log |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue |
User |
|
|
|
|
TABLE |
| CollectionEventParameterValue_log |
|
|
|
|
|
TABLE |
| CollectionEventProperty |
User |
|
|
|
|
TABLE |
| CollectionEventProperty_log |
|
|
|
|
|
TABLE |
| CollectionEventRegulation |
User |
|
|
|
|
TABLE |
| CollectionEventRegulation_log |
|
|
|
|
|
TABLE |
| CollectionEventSeries |
User |
|
|
|
|
TABLE |
| CollectionEventSeries_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
User |
|
|
|
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
User |
|
|
|
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
User |
|
|
|
|
TABLE |
| CollectionImage_log |
|
|
|
|
|
TABLE |
| CollectionManager |
|
|
|
|
|
TABLE |
| CollectionProject |
User |
|
|
|
|
TABLE |
| CollectionProject_log |
|
|
|
|
|
TABLE |
| CollectionSpecimen |
User |
|
|
|
|
TABLE |
| CollectionSpecimen_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage |
User |
|
|
|
|
TABLE |
| CollectionSpecimenImage_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
User |
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart |
User |
|
|
|
|
TABLE |
| CollectionSpecimenPart_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
User |
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing |
User |
|
|
|
|
TABLE |
| CollectionSpecimenProcessing_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
User |
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
User |
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
User |
|
|
|
|
TABLE |
| CollectionSpecimenReference_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation |
User |
|
|
|
|
TABLE |
| CollectionSpecimenRelation_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction |
User |
|
|
|
|
TABLE |
| CollectionSpecimenTransaction_log |
|
|
|
|
|
TABLE |
| CollectionTask |
User |
|
|
|
|
TABLE |
| CollectionTask_log |
|
|
|
|
|
TABLE |
| CollectionTaskImage |
User |
|
|
|
|
TABLE |
| CollectionTaskImage_log |
|
|
|
|
|
TABLE |
| CollectionTaskMetric |
User |
|
|
|
|
TABLE |
| CollectionTaskMetric_log |
|
|
|
|
|
TABLE |
| CollEventDateCategory_Enum |
User |
|
|
|
|
TABLE |
| CollEventImageType_Enum |
User |
|
|
|
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
User |
|
|
|
|
TABLE |
| CollExchangeType_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationCategory_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationDateCategory_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationQualifier_Enum |
User |
|
|
|
|
TABLE |
| CollLabelTranscriptionState_Enum |
User |
|
|
|
|
TABLE |
| CollLabelType_Enum |
User |
|
|
|
|
TABLE |
| CollMaterialCategory_Enum |
User |
|
|
|
|
TABLE |
| CollRetrievalType_Enum |
User |
|
|
|
|
TABLE |
| CollSpecimenImageType_Enum |
User |
|
|
|
|
TABLE |
| CollSpecimenRelationType_Enum |
User |
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
User |
|
|
|
|
TABLE |
| CollTaxonomicGroup_Enum |
User |
|
|
|
|
TABLE |
| CollTransactionType_Enum |
User |
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
User |
|
|
|
|
TABLE |
| CollUnitRelationType_Enum |
User |
|
|
|
|
TABLE |
| Entity |
User |
|
|
|
|
TABLE |
| EntityAccessibility_Enum |
User |
|
|
|
|
TABLE |
| EntityContext_Enum |
User |
|
|
|
|
TABLE |
| EntityDetermination_Enum |
User |
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
User |
|
|
|
|
TABLE |
| EntityRepresentation |
User |
|
|
|
|
TABLE |
| EntityUsage |
User |
|
|
|
|
TABLE |
| EntityUsage_Enum |
User |
|
|
|
|
TABLE |
| EntityVisibility_Enum |
User |
|
|
|
|
TABLE |
| ExternalIdentifier |
User |
|
|
|
|
TABLE |
| ExternalIdentifier_log |
|
|
|
|
|
TABLE |
| ExternalIdentifierType |
User |
|
|
|
|
TABLE |
| ExternalIdentifierType_log |
|
|
|
|
|
TABLE |
| Identification |
User |
|
|
|
|
TABLE |
| Identification_log |
|
|
|
|
|
TABLE |
| IdentificationUnit |
User |
|
|
|
|
TABLE |
| IdentificationUnit_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis |
User |
|
|
|
|
TABLE |
| IdentificationUnitAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
User |
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
User |
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis |
User |
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart |
User |
|
|
|
|
TABLE |
| IdentificationUnitInPart_log |
|
|
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
User |
|
|
|
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
User |
|
|
|
|
TABLE |
| Method_log |
|
|
|
|
|
TABLE |
| MethodForAnalysis |
User |
|
|
|
|
TABLE |
| MethodForProcessing |
User |
|
|
|
|
TABLE |
| Parameter |
User |
|
|
|
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
User |
|
|
|
|
TABLE |
| Processing |
User |
|
|
|
|
TABLE |
| Processing_log |
|
|
|
|
|
TABLE |
| ProcessingMaterialCategory |
User |
|
|
|
|
TABLE |
| ProjectAnalysis |
User |
|
|
|
|
TABLE |
| ProjectMaterialCategory |
User |
|
|
|
|
TABLE |
| ProjectProcessing |
User |
|
|
|
|
TABLE |
| ProjectTaxonomicGroup |
User |
|
|
|
|
TABLE |
| ProjectUser |
User |
|
|
|
|
TABLE |
| Property |
User |
|
|
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
User |
|
|
|
|
TABLE |
| ReplicationPublisher |
|
|
|
|
|
TABLE |
| Task |
User |
|
|
|
|
TABLE |
| Task_log |
|
|
|
|
|
TABLE |
| TaskDateType_Enum |
User |
|
|
|
|
TABLE |
| TaskModule |
User |
|
|
|
|
TABLE |
| TaskModule_log |
|
|
|
|
|
TABLE |
| TaskModuleType_Enum |
User |
|
|
|
|
TABLE |
| TaskResult |
User |
|
|
|
|
TABLE |
| TaskResult_log |
|
|
|
|
|
TABLE |
| TaskType_Enum |
User |
|
|
|
|
TABLE |
| Transaction |
User |
|
|
|
|
TABLE |
| Transaction_log |
|
|
|
|
|
TABLE |
| TransactionAgent |
|
|
|
|
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
User |
|
|
|
|
TABLE |
| TransactionDocument |
User |
|
|
|
|
TABLE |
| TransactionDocument_log |
|
|
|
|
|
TABLE |
| TransactionPayment |
|
|
|
|
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
User |
|
|
|
|
VIEW |
| AnnotationPart |
User |
|
|
|
|
VIEW |
| AnnotationSpecimen |
User |
|
|
|
|
VIEW |
| AnnotationUnit |
User |
|
|
|
|
VIEW |
| CollectionAgent_Core |
User |
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
User |
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
User |
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
User |
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
|
|
|
|
|
VIEW |
| FirstLinesIdentification |
User |
|
|
|
|
VIEW |
| Identification_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
User |
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
User |
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
User |
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
User |
|
|
|
|
VIEW |
| TransactionList_H7 |
User |
|
|
|
|
VIEW |
| TransactionPermit |
User |
|
|
|
|
VIEW |
| TransactionRegulation |
User |
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
User |
|
|
|
|
VIEW |
| ViewCollectionEventImage |
User |
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
User |
|
|
|
|
VIEW |
| ViewCollectionImage |
User |
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
User |
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
User |
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
User |
|
|
|
|
VIEW |
| AgentOneString |
|
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
User |
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
User |
|
|
|
|
FUNCTION |
| AnalysisList |
User |
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
User |
|
|
|
|
FUNCTION |
| AnalysisProjectList |
User |
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
User |
FUNCTION |
| CollCharacterType_List |
User |
|
|
|
|
FUNCTION |
| CollDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollectionChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
User |
|
|
|
|
FUNCTION |
| CollectionLocation |
User |
|
|
|
|
FUNCTION |
| CollectionLocationAll |
User |
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
User |
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
User |
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
User |
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollEventImageType_List |
User |
|
|
|
|
FUNCTION |
| CollExchangeType_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
User |
|
|
|
|
FUNCTION |
| CollLabelType_List |
User |
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
User |
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
User |
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
User |
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
User |
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
User |
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
|
|
|
|
|
FUNCTION |
| CurrentUser |
User |
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
User |
FUNCTION |
| DefaultProjectID |
|
|
|
|
User |
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
User |
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
User |
FUNCTION |
| EntityInformation_2 |
User |
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
User |
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
User |
FUNCTION |
| EventSeriesChildNodes |
User |
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
User |
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
User |
FUNCTION |
| EventSuperiorList |
User |
|
|
|
|
FUNCTION |
| FirstLines_4 |
User |
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesSeries |
User |
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
User |
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
User |
|
|
|
|
FUNCTION |
| ManagerCollectionList |
|
|
|
|
|
FUNCTION |
| MethodChildNodes |
User |
|
|
|
|
FUNCTION |
| MethodHierarchy |
User |
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
User |
|
|
|
|
FUNCTION |
| NameList |
User |
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
User |
|
|
|
|
FUNCTION |
| NameListPlants |
User |
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
|
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
|
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
User |
FUNCTION |
| ProcessingChildNodes |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
User |
|
|
|
|
FUNCTION |
| ProcessingListForPart |
User |
|
|
|
|
FUNCTION |
| ProcessingProjectList |
User |
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
User |
FUNCTION |
| RequesterCollectionList |
|
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
User |
FUNCTION |
| TaskChildNodes |
User |
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaskHierarchy |
User |
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
User |
FUNCTION |
| TransactionChildNodes |
User |
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
User |
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
User |
FUNCTION |
| TransactionHierarchy |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| UserCollectionList |
User |
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
User |
FUNCTION |
| Version |
|
|
|
|
User |
FUNCTION |
| VersionClient |
|
|
|
|
User |
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
User |
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
|
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
User |
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
|
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| SetXmlValue |
|
|
|
|
User |
PROCEDURE |
| Inheriting from roles: |
|
|
|
|
|
|
Role CacheUser
Reading access to objects related to the cache database
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
User |
|
|
|
|
TABLE |
| Analysis_log |
|
|
|
|
|
TABLE |
| AnalysisResult |
User |
|
|
|
|
TABLE |
| AnalysisTaxonomicGroup |
User |
|
|
|
|
TABLE |
| Annotation |
User |
User |
User |
|
|
TABLE |
| Annotation_log |
|
|
|
|
|
TABLE |
| AnnotationType_Enum |
User |
|
|
|
|
TABLE |
| AnonymCollector |
• |
|
|
|
|
TABLE |
| CacheDatabase2 |
• |
|
|
|
|
TABLE |
| CacheDescription |
User |
User |
User |
User |
|
TABLE |
| CollCircumstances_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionType_Enum |
User |
|
|
|
|
TABLE |
| CollDateCategory_Enum |
User |
|
|
|
|
TABLE |
| Collection |
User |
|
|
|
|
TABLE |
| Collection_log |
|
|
|
|
|
TABLE |
| CollectionAgent |
User |
|
|
|
|
TABLE |
| CollectionAgent_log |
|
|
|
|
|
TABLE |
| CollectionEvent |
User |
|
|
|
|
TABLE |
| CollectionEvent_log |
|
|
|
|
|
TABLE |
| CollectionEventImage |
User |
|
|
|
|
TABLE |
| CollectionEventImage_log |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation |
User |
|
|
|
|
TABLE |
| CollectionEventLocalisation_log |
|
|
|
|
|
TABLE |
| CollectionEventMethod |
User |
|
|
|
|
TABLE |
| CollectionEventMethod_log |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue |
User |
|
|
|
|
TABLE |
| CollectionEventParameterValue_log |
|
|
|
|
|
TABLE |
| CollectionEventProperty |
User |
|
|
|
|
TABLE |
| CollectionEventProperty_log |
|
|
|
|
|
TABLE |
| CollectionEventRegulation |
User |
|
|
|
|
TABLE |
| CollectionEventRegulation_log |
|
|
|
|
|
TABLE |
| CollectionEventSeries |
User |
|
|
|
|
TABLE |
| CollectionEventSeries_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
User |
|
|
|
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
User |
|
|
|
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
User |
|
|
|
|
TABLE |
| CollectionImage_log |
|
|
|
|
|
TABLE |
| CollectionManager |
|
|
|
|
|
TABLE |
| CollectionProject |
User |
|
|
|
|
TABLE |
| CollectionProject_log |
|
|
|
|
|
TABLE |
| CollectionSpecimen |
User |
|
|
|
|
TABLE |
| CollectionSpecimen_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage |
User |
|
|
|
|
TABLE |
| CollectionSpecimenImage_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
User |
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart |
User |
|
|
|
|
TABLE |
| CollectionSpecimenPart_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
User |
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing |
User |
|
|
|
|
TABLE |
| CollectionSpecimenProcessing_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
User |
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
User |
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
User |
|
|
|
|
TABLE |
| CollectionSpecimenReference_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation |
User |
|
|
|
|
TABLE |
| CollectionSpecimenRelation_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction |
User |
|
|
|
|
TABLE |
| CollectionSpecimenTransaction_log |
|
|
|
|
|
TABLE |
| CollectionTask |
User |
|
|
|
|
TABLE |
| CollectionTask_log |
|
|
|
|
|
TABLE |
| CollectionTaskImage |
User |
|
|
|
|
TABLE |
| CollectionTaskImage_log |
|
|
|
|
|
TABLE |
| CollectionTaskMetric |
User |
|
|
|
|
TABLE |
| CollectionTaskMetric_log |
|
|
|
|
|
TABLE |
| CollEventDateCategory_Enum |
User |
|
|
|
|
TABLE |
| CollEventImageType_Enum |
User |
|
|
|
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
User |
|
|
|
|
TABLE |
| CollExchangeType_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationCategory_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationDateCategory_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationQualifier_Enum |
User |
|
|
|
|
TABLE |
| CollLabelTranscriptionState_Enum |
User |
|
|
|
|
TABLE |
| CollLabelType_Enum |
User |
|
|
|
|
TABLE |
| CollMaterialCategory_Enum |
User |
|
|
|
|
TABLE |
| CollRetrievalType_Enum |
User |
|
|
|
|
TABLE |
| CollSpecimenImageType_Enum |
User |
|
|
|
|
TABLE |
| CollSpecimenRelationType_Enum |
User |
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
User |
|
|
|
|
TABLE |
| CollTaxonomicGroup_Enum |
User |
|
|
|
|
TABLE |
| CollTransactionType_Enum |
User |
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
User |
|
|
|
|
TABLE |
| CollUnitRelationType_Enum |
User |
|
|
|
|
TABLE |
| Entity |
User |
|
|
|
|
TABLE |
| EntityAccessibility_Enum |
User |
|
|
|
|
TABLE |
| EntityContext_Enum |
User |
|
|
|
|
TABLE |
| EntityDetermination_Enum |
User |
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
User |
|
|
|
|
TABLE |
| EntityRepresentation |
User |
|
|
|
|
TABLE |
| EntityUsage |
User |
|
|
|
|
TABLE |
| EntityUsage_Enum |
User |
|
|
|
|
TABLE |
| EntityVisibility_Enum |
User |
|
|
|
|
TABLE |
| ExternalIdentifier |
User |
|
|
|
|
TABLE |
| ExternalIdentifier_log |
|
|
|
|
|
TABLE |
| ExternalIdentifierType |
User |
|
|
|
|
TABLE |
| ExternalIdentifierType_log |
|
|
|
|
|
TABLE |
| Identification |
User |
|
|
|
|
TABLE |
| Identification_log |
|
|
|
|
|
TABLE |
| IdentificationUnit |
User |
|
|
|
|
TABLE |
| IdentificationUnit_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis |
User |
|
|
|
|
TABLE |
| IdentificationUnitAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
User |
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
User |
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis |
User |
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart |
User |
|
|
|
|
TABLE |
| IdentificationUnitInPart_log |
|
|
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
User |
|
|
|
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
User |
|
|
|
|
TABLE |
| Method_log |
|
|
|
|
|
TABLE |
| MethodForAnalysis |
User |
|
|
|
|
TABLE |
| MethodForProcessing |
User |
|
|
|
|
TABLE |
| Parameter |
User |
|
|
|
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
User |
|
|
|
|
TABLE |
| Processing |
User |
|
|
|
|
TABLE |
| Processing_log |
|
|
|
|
|
TABLE |
| ProcessingMaterialCategory |
User |
|
|
|
|
TABLE |
| ProjectAnalysis |
User |
|
|
|
|
TABLE |
| ProjectMaterialCategory |
User |
|
|
|
|
TABLE |
| ProjectProcessing |
User |
|
|
|
|
TABLE |
| ProjectTaxonomicGroup |
User |
|
|
|
|
TABLE |
| ProjectUser |
User |
|
|
|
|
TABLE |
| Property |
User |
|
|
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
User |
|
|
|
|
TABLE |
| ReplicationPublisher |
|
|
|
|
|
TABLE |
| Task |
User |
|
|
|
|
TABLE |
| Task_log |
|
|
|
|
|
TABLE |
| TaskDateType_Enum |
User |
|
|
|
|
TABLE |
| TaskModule |
User |
|
|
|
|
TABLE |
| TaskModule_log |
|
|
|
|
|
TABLE |
| TaskModuleType_Enum |
User |
|
|
|
|
TABLE |
| TaskResult |
User |
|
|
|
|
TABLE |
| TaskResult_log |
|
|
|
|
|
TABLE |
| TaskType_Enum |
User |
|
|
|
|
TABLE |
| Transaction |
User |
|
|
|
|
TABLE |
| Transaction_log |
|
|
|
|
|
TABLE |
| TransactionAgent |
|
|
|
|
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
User |
|
|
|
|
TABLE |
| TransactionDocument |
User |
|
|
|
|
TABLE |
| TransactionDocument_log |
|
|
|
|
|
TABLE |
| TransactionPayment |
|
|
|
|
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
User |
|
|
|
|
VIEW |
| AnnotationPart |
User |
|
|
|
|
VIEW |
| AnnotationSpecimen |
User |
|
|
|
|
VIEW |
| AnnotationUnit |
User |
|
|
|
|
VIEW |
| CollectionAgent_Core |
User |
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
User |
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
User |
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
User |
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
|
|
|
|
|
VIEW |
| FirstLinesIdentification |
User |
|
|
|
|
VIEW |
| Identification_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
User |
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
User |
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
User |
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
User |
|
|
|
|
VIEW |
| TransactionList_H7 |
User |
|
|
|
|
VIEW |
| TransactionPermit |
User |
|
|
|
|
VIEW |
| TransactionRegulation |
User |
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
User |
|
|
|
|
VIEW |
| ViewCollectionEventImage |
User |
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
User |
|
|
|
|
VIEW |
| ViewCollectionImage |
User |
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
User |
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
User |
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
User |
|
|
|
|
VIEW |
| AgentOneString |
|
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
User |
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
User |
|
|
|
|
FUNCTION |
| AnalysisList |
User |
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
User |
|
|
|
|
FUNCTION |
| AnalysisProjectList |
User |
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
User |
FUNCTION |
| CollCharacterType_List |
User |
|
|
|
|
FUNCTION |
| CollDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollectionChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
User |
|
|
|
|
FUNCTION |
| CollectionLocation |
User |
|
|
|
|
FUNCTION |
| CollectionLocationAll |
User |
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
User |
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
User |
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
User |
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollEventImageType_List |
User |
|
|
|
|
FUNCTION |
| CollExchangeType_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
User |
|
|
|
|
FUNCTION |
| CollLabelType_List |
User |
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
User |
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
User |
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
User |
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
User |
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
User |
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
|
|
|
|
|
FUNCTION |
| CurrentUser |
User |
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
User |
FUNCTION |
| DefaultProjectID |
|
|
|
|
User |
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
User |
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
User |
FUNCTION |
| EntityInformation_2 |
User |
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
User |
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
User |
FUNCTION |
| EventSeriesChildNodes |
User |
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
User |
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
User |
FUNCTION |
| EventSuperiorList |
User |
|
|
|
|
FUNCTION |
| FirstLines_4 |
User |
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesSeries |
User |
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
User |
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
User |
|
|
|
|
FUNCTION |
| ManagerCollectionList |
|
|
|
|
|
FUNCTION |
| MethodChildNodes |
User |
|
|
|
|
FUNCTION |
| MethodHierarchy |
User |
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
User |
|
|
|
|
FUNCTION |
| NameList |
User |
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
User |
|
|
|
|
FUNCTION |
| NameListPlants |
User |
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
|
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
|
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
User |
FUNCTION |
| ProcessingChildNodes |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
User |
|
|
|
|
FUNCTION |
| ProcessingListForPart |
User |
|
|
|
|
FUNCTION |
| ProcessingProjectList |
User |
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
User |
FUNCTION |
| RequesterCollectionList |
|
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
User |
FUNCTION |
| TaskChildNodes |
User |
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaskHierarchy |
User |
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
User |
FUNCTION |
| TransactionChildNodes |
User |
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
User |
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
User |
FUNCTION |
| TransactionHierarchy |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| UserCollectionList |
User |
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
User |
FUNCTION |
| Version |
|
|
|
|
User |
FUNCTION |
| VersionClient |
|
|
|
|
User |
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
User |
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
|
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
User |
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
|
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| SetXmlValue |
|
|
|
|
User |
PROCEDURE |
| Inheriting from roles: |
|
|
|
|
|
|
Role CollectionManager
Role for the administration of collections and References
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
User |
|
|
|
|
TABLE |
| Analysis_log |
|
|
|
|
|
TABLE |
| AnalysisResult |
User |
|
|
|
|
TABLE |
| AnalysisTaxonomicGroup |
User |
|
|
|
|
TABLE |
| Annotation |
User |
User |
User |
|
|
TABLE |
| Annotation_log |
|
|
|
|
|
TABLE |
| AnnotationType_Enum |
User |
|
|
|
|
TABLE |
| AnonymCollector |
|
|
|
|
|
TABLE |
| CacheDatabase2 |
|
|
|
|
|
TABLE |
| CacheDescription |
User |
User |
User |
User |
|
TABLE |
| CollCircumstances_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionType_Enum |
User |
|
|
|
|
TABLE |
| CollDateCategory_Enum |
User |
|
|
|
|
TABLE |
| Collection |
• |
• |
• |
|
|
TABLE |
| Collection_log |
|
• |
|
|
|
TABLE |
| CollectionAgent |
User |
|
|
|
|
TABLE |
| CollectionAgent_log |
|
|
|
|
|
TABLE |
| CollectionEvent |
User |
|
|
|
|
TABLE |
| CollectionEvent_log |
|
|
|
|
|
TABLE |
| CollectionEventImage |
User |
|
|
|
|
TABLE |
| CollectionEventImage_log |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation |
User |
|
|
|
|
TABLE |
| CollectionEventLocalisation_log |
|
|
|
|
|
TABLE |
| CollectionEventMethod |
User |
|
|
|
|
TABLE |
| CollectionEventMethod_log |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue |
User |
|
|
|
|
TABLE |
| CollectionEventParameterValue_log |
|
|
|
|
|
TABLE |
| CollectionEventProperty |
User |
|
|
|
|
TABLE |
| CollectionEventProperty_log |
|
|
|
|
|
TABLE |
| CollectionEventRegulation |
User |
|
|
|
|
TABLE |
| CollectionEventRegulation_log |
|
|
|
|
|
TABLE |
| CollectionEventSeries |
User |
|
|
|
|
TABLE |
| CollectionEventSeries_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
User |
|
|
|
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
User |
|
|
|
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
• |
• |
• |
• |
|
TABLE |
| CollectionImage_log |
• |
• |
|
|
|
TABLE |
| CollectionManager |
• |
• |
|
|
|
TABLE |
| CollectionProject |
User |
|
|
|
|
TABLE |
| CollectionProject_log |
|
|
|
|
|
TABLE |
| CollectionSpecimen |
User |
|
|
• |
|
TABLE |
| CollectionSpecimen_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage |
User |
|
|
|
|
TABLE |
| CollectionSpecimenImage_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
User |
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart |
User |
Storage Manager |
Storage Manager |
Storage Manager |
|
TABLE |
| CollectionSpecimenPart_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
User |
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing |
User |
Storage Manager |
Storage Manager |
Storage Manager |
|
TABLE |
| CollectionSpecimenProcessing_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
User |
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
User |
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
User |
|
|
|
|
TABLE |
| CollectionSpecimenReference_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation |
User |
|
|
|
|
TABLE |
| CollectionSpecimenRelation_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction |
• |
• |
• |
|
|
TABLE |
| CollectionSpecimenTransaction_log |
|
• |
|
|
|
TABLE |
| CollectionTask |
User |
• |
• |
• |
|
TABLE |
| CollectionTask_log |
• |
• |
|
|
|
TABLE |
| CollectionTaskImage |
User |
• |
• |
• |
|
TABLE |
| CollectionTaskImage_log |
• |
• |
|
|
|
TABLE |
| CollectionTaskMetric |
User |
• |
• |
• |
|
TABLE |
| CollectionTaskMetric_log |
• |
• |
|
|
|
TABLE |
| CollEventDateCategory_Enum |
User |
|
|
|
|
TABLE |
| CollEventImageType_Enum |
User |
|
|
|
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
User |
|
|
|
|
TABLE |
| CollExchangeType_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationCategory_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationDateCategory_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationQualifier_Enum |
User |
|
|
|
|
TABLE |
| CollLabelTranscriptionState_Enum |
User |
|
|
|
|
TABLE |
| CollLabelType_Enum |
User |
|
|
|
|
TABLE |
| CollMaterialCategory_Enum |
User |
|
|
|
|
TABLE |
| CollRetrievalType_Enum |
User |
|
|
|
|
TABLE |
| CollSpecimenImageType_Enum |
User |
|
|
|
|
TABLE |
| CollSpecimenRelationType_Enum |
User |
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
User |
|
|
|
|
TABLE |
| CollTaxonomicGroup_Enum |
User |
|
|
|
|
TABLE |
| CollTransactionType_Enum |
User |
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
User |
|
|
|
|
TABLE |
| CollUnitRelationType_Enum |
User |
|
|
|
|
TABLE |
| Entity |
User |
|
|
|
|
TABLE |
| EntityAccessibility_Enum |
User |
|
|
|
|
TABLE |
| EntityContext_Enum |
User |
|
|
|
|
TABLE |
| EntityDetermination_Enum |
User |
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
User |
|
|
|
|
TABLE |
| EntityRepresentation |
User |
|
|
|
|
TABLE |
| EntityUsage |
User |
|
|
|
|
TABLE |
| EntityUsage_Enum |
User |
|
|
|
|
TABLE |
| EntityVisibility_Enum |
User |
|
|
|
|
TABLE |
| ExternalIdentifier |
User |
|
|
|
|
TABLE |
| ExternalIdentifier_log |
|
|
|
|
|
TABLE |
| ExternalIdentifierType |
User |
|
|
|
|
TABLE |
| ExternalIdentifierType_log |
|
|
|
|
|
TABLE |
| Identification |
User |
|
|
|
|
TABLE |
| Identification_log |
|
|
|
|
|
TABLE |
| IdentificationUnit |
User |
|
|
|
|
TABLE |
| IdentificationUnit_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis |
User |
Storage Manager |
Storage Manager |
Storage Manager |
|
TABLE |
| IdentificationUnitAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
User |
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
User |
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis |
User |
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart |
User |
|
Storage Manager |
Storage Manager |
|
TABLE |
| IdentificationUnitInPart_log |
|
|
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
User |
|
|
|
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
User |
|
|
|
|
TABLE |
| Method_log |
|
|
|
|
|
TABLE |
| MethodForAnalysis |
User |
|
|
|
|
TABLE |
| MethodForProcessing |
User |
|
|
|
|
TABLE |
| Parameter |
User |
|
|
|
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
User |
|
|
|
|
TABLE |
| Processing |
User |
|
|
|
|
TABLE |
| Processing_log |
|
|
|
|
|
TABLE |
| ProcessingMaterialCategory |
User |
|
|
|
|
TABLE |
| ProjectAnalysis |
User |
|
|
|
|
TABLE |
| ProjectMaterialCategory |
User |
|
|
|
|
TABLE |
| ProjectProcessing |
User |
|
|
|
|
TABLE |
| ProjectTaxonomicGroup |
User |
|
|
|
|
TABLE |
| ProjectUser |
User |
|
|
|
|
TABLE |
| Property |
User |
|
|
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
User |
|
|
|
|
TABLE |
| ReplicationPublisher |
|
|
|
|
|
TABLE |
| Task |
User |
• |
• |
|
|
TABLE |
| Task_log |
• |
• |
|
|
|
TABLE |
| TaskDateType_Enum |
User |
|
|
|
|
TABLE |
| TaskModule |
User |
• |
• |
• |
|
TABLE |
| TaskModule_log |
• |
• |
|
|
|
TABLE |
| TaskModuleType_Enum |
User |
|
|
|
|
TABLE |
| TaskResult |
User |
• |
• |
• |
|
TABLE |
| TaskResult_log |
• |
• |
|
|
|
TABLE |
| TaskType_Enum |
User |
|
|
|
|
TABLE |
| Transaction |
• |
• |
• |
• |
|
TABLE |
| Transaction_log |
|
• |
|
|
|
TABLE |
| TransactionAgent |
Transaction User |
• |
• |
• |
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
User |
|
|
|
|
TABLE |
| TransactionDocument |
• |
• |
• |
• |
|
TABLE |
| TransactionDocument_log |
|
• |
|
|
|
TABLE |
| TransactionPayment |
Transaction User |
• |
• |
• |
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
User |
|
|
|
|
VIEW |
| AnnotationPart |
User |
|
|
|
|
VIEW |
| AnnotationSpecimen |
User |
|
|
|
|
VIEW |
| AnnotationUnit |
User |
|
|
|
|
VIEW |
| CollectionAgent_Core |
User |
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
User |
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
User |
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
User |
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
|
|
|
|
|
VIEW |
| FirstLinesIdentification |
User |
|
|
|
|
VIEW |
| Identification_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
User |
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
User |
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
User |
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
User |
|
|
|
|
VIEW |
| TransactionList_H7 |
User |
|
|
|
|
VIEW |
| TransactionPermit |
User |
|
|
|
|
VIEW |
| TransactionRegulation |
User |
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
User |
|
|
|
|
VIEW |
| ViewCollectionEventImage |
User |
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
User |
|
|
|
|
VIEW |
| ViewCollectionImage |
User |
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
User |
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
User |
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
User |
|
|
|
|
VIEW |
| AgentOneString |
|
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
User |
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
User |
|
|
|
|
FUNCTION |
| AnalysisList |
User |
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
User |
|
|
|
|
FUNCTION |
| AnalysisProjectList |
User |
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
User |
FUNCTION |
| CollCharacterType_List |
User |
|
|
|
|
FUNCTION |
| CollDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollectionChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
User |
|
|
|
|
FUNCTION |
| CollectionLocation |
User |
|
|
|
|
FUNCTION |
| CollectionLocationAll |
User |
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
User |
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
User |
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
User |
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollEventImageType_List |
User |
|
|
|
|
FUNCTION |
| CollExchangeType_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
User |
|
|
|
|
FUNCTION |
| CollLabelType_List |
User |
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
User |
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
User |
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
User |
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
User |
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
User |
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
• |
|
|
|
|
FUNCTION |
| CurrentUser |
User |
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
User |
FUNCTION |
| DefaultProjectID |
|
|
|
|
User |
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
User |
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
User |
FUNCTION |
| EntityInformation_2 |
User |
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
User |
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
User |
FUNCTION |
| EventSeriesChildNodes |
User |
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
User |
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
User |
FUNCTION |
| EventSuperiorList |
User |
|
|
|
|
FUNCTION |
| FirstLines_4 |
User |
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesSeries |
User |
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
User |
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
User |
|
|
|
|
FUNCTION |
| ManagerCollectionList |
• |
|
|
|
|
FUNCTION |
| MethodChildNodes |
User |
|
|
|
|
FUNCTION |
| MethodHierarchy |
User |
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
User |
|
|
|
|
FUNCTION |
| NameList |
User |
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
User |
|
|
|
|
FUNCTION |
| NameListPlants |
User |
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
|
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
|
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
User |
FUNCTION |
| ProcessingChildNodes |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
User |
|
|
|
|
FUNCTION |
| ProcessingListForPart |
User |
|
|
|
|
FUNCTION |
| ProcessingProjectList |
User |
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
User |
FUNCTION |
| RequesterCollectionList |
|
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
User |
FUNCTION |
| TaskChildNodes |
User |
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaskHierarchy |
User |
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
User |
FUNCTION |
| TransactionChildNodes |
User |
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
User |
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
User |
FUNCTION |
| TransactionHierarchy |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| UserCollectionList |
User |
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
User |
FUNCTION |
| Version |
|
|
|
|
User |
FUNCTION |
| VersionClient |
|
|
|
|
User |
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
User |
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
|
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
User |
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
|
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| SetXmlValue |
|
|
|
|
User |
PROCEDURE |
| Inheriting from roles: |
|
|
|
|
|
|
- StorageManager
- TransactionUser
- User
Role DataManager
Can insert, update and delete data from tables Analysis, Annotation, CollectionEvent, CollectionSpecimen, Method, MethodForAnalysis, MethodForProcessing, Processing, ProcessingMaterialCategory, ProjectAnalysis, ProjectProcessing
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
Editor |
• |
• |
• |
|
TABLE |
| Analysis_log |
|
• |
|
|
|
TABLE |
| AnalysisResult |
User |
• |
• |
• |
|
TABLE |
| AnalysisTaxonomicGroup |
User |
• |
• |
• |
|
TABLE |
| Annotation |
User |
User |
• |
• |
|
TABLE |
| Annotation_log |
Editor |
Editor |
|
|
|
TABLE |
| AnnotationType_Enum |
User |
|
|
|
|
TABLE |
| AnonymCollector |
|
|
|
|
|
TABLE |
| CacheDatabase2 |
|
|
|
|
|
TABLE |
| CacheDescription |
User |
User |
User |
User |
|
TABLE |
| CollCircumstances_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionType_Enum |
User |
|
|
|
|
TABLE |
| CollDateCategory_Enum |
User |
|
|
|
|
TABLE |
| Collection |
Editor |
|
Editor |
|
|
TABLE |
| Collection_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionAgent |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionAgent_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionEvent |
User |
Editor |
Editor |
• |
|
TABLE |
| CollectionEvent_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionEventImage |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionEventImage_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionEventLocalisation |
User |
Typist |
Typist |
Editor |
|
TABLE |
| CollectionEventLocalisation_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionEventMethod |
User |
• |
• |
• |
|
TABLE |
| CollectionEventMethod_log |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue |
User |
• |
• |
• |
|
TABLE |
| CollectionEventParameterValue_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionEventProperty |
User |
Typist |
Typist |
Editor |
|
TABLE |
| CollectionEventProperty_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionEventRegulation |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionEventRegulation_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionEventSeries |
User |
Editor |
Typist |
Editor |
|
TABLE |
| CollectionEventSeries_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
User |
Editor |
Typist |
Editor |
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
User |
• |
• |
|
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
User |
Editor |
Editor |
|
|
TABLE |
| CollectionImage_log |
|
|
|
|
|
TABLE |
| CollectionManager |
|
|
|
|
|
TABLE |
| CollectionProject |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionProject_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimen |
Typist |
Editor |
Editor |
• |
|
TABLE |
| CollectionSpecimen_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimenImage |
Typist |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenImage_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenImageProperty_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimenPart |
User |
Typist |
Typist |
Editor |
|
TABLE |
| CollectionSpecimenPart_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenPartDescription_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimenProcessing |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenProcessing_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenReference_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimenRelation |
User |
Editor |
Editor |
Editor |
|
TABLE |
| CollectionSpecimenRelation_log |
Editor |
Editor |
|
|
|
TABLE |
| CollectionSpecimenTransaction |
User |
Typist |
Typist |
Editor |
|
TABLE |
| CollectionSpecimenTransaction_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionTask |
User |
|
|
|
|
TABLE |
| CollectionTask_log |
|
|
|
|
|
TABLE |
| CollectionTaskImage |
User |
|
|
|
|
TABLE |
| CollectionTaskImage_log |
|
|
|
|
|
TABLE |
| CollectionTaskMetric |
User |
|
|
|
|
TABLE |
| CollectionTaskMetric_log |
|
|
|
|
|
TABLE |
| CollEventDateCategory_Enum |
Editor |
|
|
|
|
TABLE |
| CollEventImageType_Enum |
Editor |
|
|
|
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
User |
|
|
|
|
TABLE |
| CollExchangeType_Enum |
Editor |
|
|
|
|
TABLE |
| CollIdentificationCategory_Enum |
Editor |
|
|
|
|
TABLE |
| CollIdentificationDateCategory_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationQualifier_Enum |
User |
|
|
|
|
TABLE |
| CollLabelTranscriptionState_Enum |
User |
|
|
|
|
TABLE |
| CollLabelType_Enum |
Editor |
|
|
|
|
TABLE |
| CollMaterialCategory_Enum |
User |
|
|
|
|
TABLE |
| CollRetrievalType_Enum |
User |
|
|
|
|
TABLE |
| CollSpecimenImageType_Enum |
User |
|
|
|
|
TABLE |
| CollSpecimenRelationType_Enum |
User |
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
User |
|
|
|
|
TABLE |
| CollTaxonomicGroup_Enum |
User |
|
|
|
|
TABLE |
| CollTransactionType_Enum |
User |
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
User |
|
|
|
|
TABLE |
| CollUnitRelationType_Enum |
User |
|
|
|
|
TABLE |
| Entity |
User |
|
|
|
|
TABLE |
| EntityAccessibility_Enum |
User |
|
|
|
|
TABLE |
| EntityContext_Enum |
User |
|
|
|
|
TABLE |
| EntityDetermination_Enum |
User |
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
User |
|
|
|
|
TABLE |
| EntityRepresentation |
User |
|
|
|
|
TABLE |
| EntityUsage |
User |
|
|
|
|
TABLE |
| EntityUsage_Enum |
User |
|
|
|
|
TABLE |
| EntityVisibility_Enum |
User |
|
|
|
|
TABLE |
| ExternalIdentifier |
User |
Editor |
Editor |
Editor |
|
TABLE |
| ExternalIdentifier_log |
Editor |
Editor |
|
|
|
TABLE |
| ExternalIdentifierType |
User |
Editor |
Editor |
Editor |
|
TABLE |
| ExternalIdentifierType_log |
Editor |
|
|
|
|
TABLE |
| Identification |
User |
Typist |
Typist |
Editor |
|
TABLE |
| Identification_log |
Editor |
Editor |
|
|
|
TABLE |
| IdentificationUnit |
User |
Editor |
Editor |
Editor |
|
TABLE |
| IdentificationUnit_log |
Editor |
Editor |
|
|
|
TABLE |
| IdentificationUnitAnalysis |
User |
Editor |
Editor |
Editor |
|
TABLE |
| IdentificationUnitAnalysis_log |
Typist |
Typist |
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
User |
Editor |
Editor |
Editor |
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
Editor |
Editor |
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
User |
Editor |
Editor |
Editor |
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
Editor |
Editor |
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis |
User |
Editor |
Editor |
Editor |
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
Editor |
|
|
|
TABLE |
| IdentificationUnitInPart |
User |
Typist |
Typist |
Editor |
|
TABLE |
| IdentificationUnitInPart_log |
Typist |
Typist |
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
User |
|
|
|
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
User |
• |
• |
• |
|
TABLE |
| Method_log |
|
• |
|
|
|
TABLE |
| MethodForAnalysis |
User |
• |
• |
• |
|
TABLE |
| MethodForProcessing |
User |
• |
• |
• |
|
TABLE |
| Parameter |
User |
• |
• |
• |
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
User |
• |
• |
• |
|
TABLE |
| Processing |
User |
• |
• |
• |
|
TABLE |
| Processing_log |
Editor |
• |
|
|
|
TABLE |
| ProcessingMaterialCategory |
User |
• |
• |
• |
|
TABLE |
| ProjectAnalysis |
User |
• |
• |
• |
|
TABLE |
| ProjectMaterialCategory |
User |
|
|
|
|
TABLE |
| ProjectProcessing |
User |
• |
• |
• |
|
TABLE |
| ProjectTaxonomicGroup |
User |
|
|
|
|
TABLE |
| ProjectUser |
User |
|
|
|
|
TABLE |
| Property |
User |
|
|
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
User |
|
|
|
|
TABLE |
| ReplicationPublisher |
Editor |
|
|
|
|
TABLE |
| Task |
User |
|
|
|
|
TABLE |
| Task_log |
|
|
|
|
|
TABLE |
| TaskDateType_Enum |
User |
|
|
|
|
TABLE |
| TaskModule |
User |
|
|
|
|
TABLE |
| TaskModule_log |
|
|
|
|
|
TABLE |
| TaskModuleType_Enum |
User |
|
|
|
|
TABLE |
| TaskResult |
User |
|
|
|
|
TABLE |
| TaskResult_log |
|
|
|
|
|
TABLE |
| TaskType_Enum |
User |
|
|
|
|
TABLE |
| Transaction |
User |
|
|
|
|
TABLE |
| Transaction_log |
Typist |
Typist |
|
|
|
TABLE |
| TransactionAgent |
|
|
|
|
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
User |
|
|
|
|
TABLE |
| TransactionDocument |
User |
Typist |
Typist |
|
|
TABLE |
| TransactionDocument_log |
Typist |
Typist |
|
|
|
TABLE |
| TransactionPayment |
|
|
|
|
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
User |
|
|
|
|
VIEW |
| AnnotationPart |
User |
|
|
|
|
VIEW |
| AnnotationSpecimen |
User |
|
|
|
|
VIEW |
| AnnotationUnit |
User |
|
|
|
|
VIEW |
| CollectionAgent_Core |
User |
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
User |
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
User |
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
User |
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
|
|
|
|
|
VIEW |
| FirstLinesIdentification |
User |
|
|
|
|
VIEW |
| Identification_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
User |
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
User |
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
User |
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
User |
|
|
|
|
VIEW |
| TransactionList_H7 |
User |
|
|
|
|
VIEW |
| TransactionPermit |
User |
|
|
|
|
VIEW |
| TransactionRegulation |
User |
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
User |
|
|
|
|
VIEW |
| ViewCollectionEventImage |
User |
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
User |
|
|
|
|
VIEW |
| ViewCollectionImage |
User |
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
User |
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
User |
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
User |
|
|
|
|
VIEW |
| AgentOneString |
Editor |
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
User |
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
User |
|
|
|
|
FUNCTION |
| AnalysisList |
User |
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
User |
|
|
|
|
FUNCTION |
| AnalysisProjectList |
User |
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
User |
FUNCTION |
| CollCharacterType_List |
Editor |
|
|
|
|
FUNCTION |
| CollDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollectionChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
User |
|
|
|
|
FUNCTION |
| CollectionLocation |
User |
|
|
|
|
FUNCTION |
| CollectionLocationAll |
User |
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
User |
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
User |
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
User |
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
Editor |
|
|
|
|
FUNCTION |
| CollEventImageType_List |
Editor |
|
|
|
|
FUNCTION |
| CollExchangeType_List |
Editor |
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
Editor |
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
User |
|
|
|
|
FUNCTION |
| CollLabelType_List |
User |
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
User |
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
User |
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
User |
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
User |
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
Editor |
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
|
|
|
|
|
FUNCTION |
| CurrentUser |
User |
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
User |
FUNCTION |
| DefaultProjectID |
|
|
|
|
User |
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
User |
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
User |
FUNCTION |
| EntityInformation_2 |
User |
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
User |
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
User |
FUNCTION |
| EventSeriesChildNodes |
User |
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
User |
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
User |
FUNCTION |
| EventSuperiorList |
User |
|
|
|
|
FUNCTION |
| FirstLines_4 |
User |
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesSeries |
User |
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
User |
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
User |
|
|
|
|
FUNCTION |
| ManagerCollectionList |
|
|
|
|
|
FUNCTION |
| MethodChildNodes |
User |
|
|
|
|
FUNCTION |
| MethodHierarchy |
User |
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
User |
|
|
|
|
FUNCTION |
| NameList |
User |
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
User |
|
|
|
|
FUNCTION |
| NameListPlants |
User |
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
Typist |
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
Editor |
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
User |
FUNCTION |
| ProcessingChildNodes |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
User |
|
|
|
|
FUNCTION |
| ProcessingListForPart |
User |
|
|
|
|
FUNCTION |
| ProcessingProjectList |
User |
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
User |
FUNCTION |
| RequesterCollectionList |
|
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
User |
FUNCTION |
| TaskChildNodes |
User |
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaskHierarchy |
User |
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
User |
FUNCTION |
| TransactionChildNodes |
User |
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
User |
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
User |
FUNCTION |
| TransactionHierarchy |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| UserCollectionList |
User |
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
User |
FUNCTION |
| Version |
|
|
|
|
User |
FUNCTION |
| VersionClient |
|
|
|
|
User |
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
User |
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
Editor |
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
Editor |
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
User |
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
Editor |
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
Editor |
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| SetXmlValue |
|
|
|
|
User |
PROCEDURE |
| Inheriting from roles: |
|
|
|
|
|
|
Role DescriptionEditor
Role with write access to the tables for the desprition of the database objects
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
|
|
|
|
|
TABLE |
| Analysis_log |
|
|
|
|
|
TABLE |
| AnalysisResult |
|
|
|
|
|
TABLE |
| AnalysisTaxonomicGroup |
|
|
|
|
|
TABLE |
| Annotation |
|
|
|
|
|
TABLE |
| Annotation_log |
|
|
|
|
|
TABLE |
| AnnotationType_Enum |
|
|
|
|
|
TABLE |
| AnonymCollector |
|
|
|
|
|
TABLE |
| CacheDatabase2 |
|
|
|
|
|
TABLE |
| CacheDescription |
|
|
|
|
|
TABLE |
| CollCircumstances_Enum |
|
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
|
|
|
|
|
TABLE |
| CollCollectionType_Enum |
|
|
|
|
|
TABLE |
| CollDateCategory_Enum |
|
|
|
|
|
TABLE |
| Collection |
|
|
|
|
|
TABLE |
| Collection_log |
|
|
|
|
|
TABLE |
| CollectionAgent |
|
|
|
|
|
TABLE |
| CollectionAgent_log |
|
|
|
|
|
TABLE |
| CollectionEvent |
|
|
|
|
|
TABLE |
| CollectionEvent_log |
|
|
|
|
|
TABLE |
| CollectionEventImage |
|
|
|
|
|
TABLE |
| CollectionEventImage_log |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation_log |
|
|
|
|
|
TABLE |
| CollectionEventMethod |
|
|
|
|
|
TABLE |
| CollectionEventMethod_log |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue_log |
|
|
|
|
|
TABLE |
| CollectionEventProperty |
|
|
|
|
|
TABLE |
| CollectionEventProperty_log |
|
|
|
|
|
TABLE |
| CollectionEventRegulation |
|
|
|
|
|
TABLE |
| CollectionEventRegulation_log |
|
|
|
|
|
TABLE |
| CollectionEventSeries |
|
|
|
|
|
TABLE |
| CollectionEventSeries_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
|
|
|
|
|
TABLE |
| CollectionImage_log |
|
|
|
|
|
TABLE |
| CollectionManager |
|
|
|
|
|
TABLE |
| CollectionProject |
|
|
|
|
|
TABLE |
| CollectionProject_log |
|
|
|
|
|
TABLE |
| CollectionSpecimen |
|
|
|
|
|
TABLE |
| CollectionSpecimen_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction_log |
|
|
|
|
|
TABLE |
| CollectionTask |
|
|
|
|
|
TABLE |
| CollectionTask_log |
|
|
|
|
|
TABLE |
| CollectionTaskImage |
|
|
|
|
|
TABLE |
| CollectionTaskImage_log |
|
|
|
|
|
TABLE |
| CollectionTaskMetric |
|
|
|
|
|
TABLE |
| CollectionTaskMetric_log |
|
|
|
|
|
TABLE |
| CollEventDateCategory_Enum |
|
|
|
|
|
TABLE |
| CollEventImageType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
|
|
|
|
|
TABLE |
| CollExchangeType_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationCategory_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationDateCategory_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationQualifier_Enum |
|
|
|
|
|
TABLE |
| CollLabelTranscriptionState_Enum |
|
|
|
|
|
TABLE |
| CollLabelType_Enum |
|
|
|
|
|
TABLE |
| CollMaterialCategory_Enum |
|
|
|
|
|
TABLE |
| CollRetrievalType_Enum |
|
|
|
|
|
TABLE |
| CollSpecimenImageType_Enum |
|
|
|
|
|
TABLE |
| CollSpecimenRelationType_Enum |
|
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
|
|
|
|
|
TABLE |
| CollTaxonomicGroup_Enum |
|
|
|
|
|
TABLE |
| CollTransactionType_Enum |
|
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
|
|
|
|
|
TABLE |
| CollUnitRelationType_Enum |
|
|
|
|
|
TABLE |
| Entity |
• |
• |
• |
• |
|
TABLE |
| EntityAccessibility_Enum |
|
|
|
|
|
TABLE |
| EntityContext_Enum |
• |
• |
• |
• |
|
TABLE |
| EntityDetermination_Enum |
|
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
|
|
|
|
|
TABLE |
| EntityRepresentation |
• |
• |
• |
• |
|
TABLE |
| EntityUsage |
• |
• |
• |
• |
|
TABLE |
| EntityUsage_Enum |
|
|
|
|
|
TABLE |
| EntityVisibility_Enum |
|
|
|
|
|
TABLE |
| ExternalIdentifier |
|
|
|
|
|
TABLE |
| ExternalIdentifier_log |
|
|
|
|
|
TABLE |
| ExternalIdentifierType |
|
|
|
|
|
TABLE |
| ExternalIdentifierType_log |
|
|
|
|
|
TABLE |
| Identification |
|
|
|
|
|
TABLE |
| Identification_log |
|
|
|
|
|
TABLE |
| IdentificationUnit |
|
|
|
|
|
TABLE |
| IdentificationUnit_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart_log |
|
|
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
|
|
|
|
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
|
|
|
|
|
TABLE |
| Method_log |
|
|
|
|
|
TABLE |
| MethodForAnalysis |
|
|
|
|
|
TABLE |
| MethodForProcessing |
|
|
|
|
|
TABLE |
| Parameter |
|
|
|
|
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
|
|
|
|
|
TABLE |
| Processing |
|
|
|
|
|
TABLE |
| Processing_log |
|
|
|
|
|
TABLE |
| ProcessingMaterialCategory |
|
|
|
|
|
TABLE |
| ProjectAnalysis |
|
|
|
|
|
TABLE |
| ProjectMaterialCategory |
|
|
|
|
|
TABLE |
| ProjectProcessing |
|
|
|
|
|
TABLE |
| ProjectTaxonomicGroup |
|
|
|
|
|
TABLE |
| ProjectUser |
|
|
|
|
|
TABLE |
| Property |
|
|
|
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
|
|
|
|
|
TABLE |
| ReplicationPublisher |
|
|
|
|
|
TABLE |
| Task |
|
|
|
|
|
TABLE |
| Task_log |
|
|
|
|
|
TABLE |
| TaskDateType_Enum |
|
|
|
|
|
TABLE |
| TaskModule |
|
|
|
|
|
TABLE |
| TaskModule_log |
|
|
|
|
|
TABLE |
| TaskModuleType_Enum |
|
|
|
|
|
TABLE |
| TaskResult |
|
|
|
|
|
TABLE |
| TaskResult_log |
|
|
|
|
|
TABLE |
| TaskType_Enum |
|
|
|
|
|
TABLE |
| Transaction |
|
|
|
|
|
TABLE |
| Transaction_log |
|
|
|
|
|
TABLE |
| TransactionAgent |
|
|
|
|
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
|
|
|
|
|
TABLE |
| TransactionDocument |
|
|
|
|
|
TABLE |
| TransactionDocument_log |
|
|
|
|
|
TABLE |
| TransactionPayment |
|
|
|
|
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
|
|
|
|
|
VIEW |
| AnnotationPart |
|
|
|
|
|
VIEW |
| AnnotationSpecimen |
|
|
|
|
|
VIEW |
| AnnotationUnit |
|
|
|
|
|
VIEW |
| CollectionAgent_Core |
|
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
|
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
|
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
|
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
|
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
|
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
|
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
|
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
|
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
|
|
|
|
|
VIEW |
| FirstLinesIdentification |
|
|
|
|
|
VIEW |
| Identification_Core2 |
|
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
|
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
|
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
|
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
|
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
|
|
|
|
|
VIEW |
| TransactionList_H7 |
|
|
|
|
|
VIEW |
| TransactionPermit |
|
|
|
|
|
VIEW |
| TransactionRegulation |
|
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
|
|
|
|
|
VIEW |
| ViewCollectionEventImage |
|
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
|
|
|
|
|
VIEW |
| ViewCollectionImage |
|
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
|
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
|
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
|
|
|
|
|
VIEW |
| AgentOneString |
|
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
|
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
|
|
|
|
|
FUNCTION |
| AnalysisList |
|
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
|
|
|
|
|
FUNCTION |
| AnalysisProjectList |
|
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
|
FUNCTION |
| CollCharacterType_List |
|
|
|
|
|
FUNCTION |
| CollDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollectionChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
|
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
|
|
|
|
|
FUNCTION |
| CollectionLocation |
|
|
|
|
|
FUNCTION |
| CollectionLocationAll |
|
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
|
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
|
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
|
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollEventImageType_List |
|
|
|
|
|
FUNCTION |
| CollExchangeType_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
|
|
|
|
|
FUNCTION |
| CollLabelType_List |
|
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
|
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
|
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
|
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
|
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
|
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
|
|
|
|
|
FUNCTION |
| CurrentUser |
|
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
|
FUNCTION |
| DefaultProjectID |
|
|
|
|
|
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
|
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
|
FUNCTION |
| EntityInformation_2 |
|
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
|
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
|
FUNCTION |
| EventSeriesChildNodes |
|
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
|
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
|
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
|
FUNCTION |
| EventSuperiorList |
|
|
|
|
|
FUNCTION |
| FirstLines_4 |
|
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
|
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
|
|
|
|
|
FUNCTION |
| FirstLinesSeries |
|
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
|
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
|
|
|
|
|
FUNCTION |
| ManagerCollectionList |
|
|
|
|
|
FUNCTION |
| MethodChildNodes |
|
|
|
|
|
FUNCTION |
| MethodHierarchy |
|
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
|
|
|
|
|
FUNCTION |
| NameList |
|
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
|
|
|
|
|
FUNCTION |
| NameListPlants |
|
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
|
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
|
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
|
FUNCTION |
| ProcessingChildNodes |
|
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
|
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
|
|
|
|
|
FUNCTION |
| ProcessingListForPart |
|
|
|
|
|
FUNCTION |
| ProcessingProjectList |
|
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
|
FUNCTION |
| RequesterCollectionList |
|
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
|
FUNCTION |
| TaskChildNodes |
|
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
|
FUNCTION |
| TaskHierarchy |
|
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
|
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
|
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
|
FUNCTION |
| TransactionChildNodes |
|
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
|
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
|
FUNCTION |
| TransactionHierarchy |
|
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
|
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
|
|
|
|
|
FUNCTION |
| UserCollectionList |
|
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
|
FUNCTION |
| Version |
|
|
|
|
|
FUNCTION |
| VersionClient |
|
|
|
|
|
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
|
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
|
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
|
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
|
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
|
PROCEDURE |
| SetXmlValue |
|
|
|
|
|
PROCEDURE |
Role Editor
Standard role for most users. Write permissions for most tables but can not delete data from tables CollectionReference and CollectionReference
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
• |
• |
• |
|
|
TABLE |
| Analysis_log |
|
|
|
|
|
TABLE |
| AnalysisResult |
User |
|
|
|
|
TABLE |
| AnalysisTaxonomicGroup |
User |
|
|
|
|
TABLE |
| Annotation |
User |
User |
User |
|
|
TABLE |
| Annotation_log |
• |
• |
|
|
|
TABLE |
| AnnotationType_Enum |
User |
|
|
|
|
TABLE |
| AnonymCollector |
|
|
|
|
|
TABLE |
| CacheDatabase2 |
|
|
|
|
|
TABLE |
| CacheDescription |
User |
User |
User |
User |
|
TABLE |
| CollCircumstances_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionType_Enum |
User |
|
|
|
|
TABLE |
| CollDateCategory_Enum |
User |
|
|
|
|
TABLE |
| Collection |
• |
|
• |
|
|
TABLE |
| Collection_log |
• |
• |
|
|
|
TABLE |
| CollectionAgent |
User |
• |
• |
• |
|
TABLE |
| CollectionAgent_log |
• |
• |
|
|
|
TABLE |
| CollectionEvent |
User |
• |
• |
|
|
TABLE |
| CollectionEvent_log |
• |
• |
|
|
|
TABLE |
| CollectionEventImage |
User |
• |
• |
• |
|
TABLE |
| CollectionEventImage_log |
• |
• |
|
|
|
TABLE |
| CollectionEventLocalisation |
User |
Typist |
Typist |
• |
|
TABLE |
| CollectionEventLocalisation_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionEventMethod |
User |
• |
• |
• |
|
TABLE |
| CollectionEventMethod_log |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue |
User |
• |
• |
• |
|
TABLE |
| CollectionEventParameterValue_log |
• |
• |
|
|
|
TABLE |
| CollectionEventProperty |
User |
Typist |
Typist |
• |
|
TABLE |
| CollectionEventProperty_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionEventRegulation |
User |
• |
• |
• |
|
TABLE |
| CollectionEventRegulation_log |
• |
• |
|
|
|
TABLE |
| CollectionEventSeries |
User |
• |
Typist |
• |
|
TABLE |
| CollectionEventSeries_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
User |
• |
Typist |
• |
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
User |
|
|
|
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
User |
• |
• |
|
|
TABLE |
| CollectionImage_log |
|
|
|
|
|
TABLE |
| CollectionManager |
|
|
|
|
|
TABLE |
| CollectionProject |
User |
• |
• |
• |
|
TABLE |
| CollectionProject_log |
• |
• |
|
|
|
TABLE |
| CollectionSpecimen |
Typist |
• |
• |
|
|
TABLE |
| CollectionSpecimen_log |
• |
• |
|
|
|
TABLE |
| CollectionSpecimenImage |
Typist |
• |
• |
• |
|
TABLE |
| CollectionSpecimenImage_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
User |
• |
• |
• |
|
TABLE |
| CollectionSpecimenImageProperty_log |
• |
• |
|
|
|
TABLE |
| CollectionSpecimenPart |
User |
Typist |
Typist |
• |
|
TABLE |
| CollectionSpecimenPart_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
User |
• |
• |
• |
|
TABLE |
| CollectionSpecimenPartDescription_log |
• |
• |
|
|
|
TABLE |
| CollectionSpecimenProcessing |
User |
• |
• |
• |
|
TABLE |
| CollectionSpecimenProcessing_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
User |
• |
• |
• |
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
• |
• |
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
User |
• |
• |
• |
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
User |
• |
• |
• |
|
TABLE |
| CollectionSpecimenReference_log |
• |
• |
|
|
|
TABLE |
| CollectionSpecimenRelation |
User |
• |
• |
• |
|
TABLE |
| CollectionSpecimenRelation_log |
• |
• |
|
|
|
TABLE |
| CollectionSpecimenTransaction |
User |
Typist |
Typist |
• |
|
TABLE |
| CollectionSpecimenTransaction_log |
Typist |
Typist |
|
|
|
TABLE |
| CollectionTask |
User |
|
|
|
|
TABLE |
| CollectionTask_log |
|
|
|
|
|
TABLE |
| CollectionTaskImage |
User |
|
|
|
|
TABLE |
| CollectionTaskImage_log |
|
|
|
|
|
TABLE |
| CollectionTaskMetric |
User |
|
|
|
|
TABLE |
| CollectionTaskMetric_log |
|
|
|
|
|
TABLE |
| CollEventDateCategory_Enum |
• |
|
|
|
|
TABLE |
| CollEventImageType_Enum |
• |
|
|
|
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
User |
|
|
|
|
TABLE |
| CollExchangeType_Enum |
• |
|
|
|
|
TABLE |
| CollIdentificationCategory_Enum |
• |
|
|
|
|
TABLE |
| CollIdentificationDateCategory_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationQualifier_Enum |
User |
|
|
|
|
TABLE |
| CollLabelTranscriptionState_Enum |
User |
|
|
|
|
TABLE |
| CollLabelType_Enum |
• |
|
|
|
|
TABLE |
| CollMaterialCategory_Enum |
User |
|
|
|
|
TABLE |
| CollRetrievalType_Enum |
User |
|
|
|
|
TABLE |
| CollSpecimenImageType_Enum |
User |
|
|
|
|
TABLE |
| CollSpecimenRelationType_Enum |
User |
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
User |
|
|
|
|
TABLE |
| CollTaxonomicGroup_Enum |
User |
|
|
|
|
TABLE |
| CollTransactionType_Enum |
User |
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
User |
|
|
|
|
TABLE |
| CollUnitRelationType_Enum |
User |
|
|
|
|
TABLE |
| Entity |
User |
|
|
|
|
TABLE |
| EntityAccessibility_Enum |
User |
|
|
|
|
TABLE |
| EntityContext_Enum |
User |
|
|
|
|
TABLE |
| EntityDetermination_Enum |
User |
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
User |
|
|
|
|
TABLE |
| EntityRepresentation |
User |
|
|
|
|
TABLE |
| EntityUsage |
User |
|
|
|
|
TABLE |
| EntityUsage_Enum |
User |
|
|
|
|
TABLE |
| EntityVisibility_Enum |
User |
|
|
|
|
TABLE |
| ExternalIdentifier |
User |
• |
• |
• |
|
TABLE |
| ExternalIdentifier_log |
• |
• |
|
|
|
TABLE |
| ExternalIdentifierType |
User |
• |
• |
• |
|
TABLE |
| ExternalIdentifierType_log |
• |
|
|
|
|
TABLE |
| Identification |
User |
Typist |
Typist |
• |
|
TABLE |
| Identification_log |
• |
• |
|
|
|
TABLE |
| IdentificationUnit |
User |
• |
• |
• |
|
TABLE |
| IdentificationUnit_log |
• |
• |
|
|
|
TABLE |
| IdentificationUnitAnalysis |
User |
• |
• |
• |
|
TABLE |
| IdentificationUnitAnalysis_log |
Typist |
Typist |
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
User |
• |
• |
• |
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
• |
• |
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
User |
• |
• |
• |
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
• |
• |
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis |
User |
• |
• |
• |
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
• |
|
|
|
TABLE |
| IdentificationUnitInPart |
User |
Typist |
Typist |
• |
|
TABLE |
| IdentificationUnitInPart_log |
Typist |
Typist |
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
User |
|
|
|
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
User |
|
|
|
|
TABLE |
| Method_log |
|
|
|
|
|
TABLE |
| MethodForAnalysis |
User |
|
|
|
|
TABLE |
| MethodForProcessing |
User |
|
|
|
|
TABLE |
| Parameter |
User |
|
|
|
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
User |
|
|
|
|
TABLE |
| Processing |
User |
• |
• |
|
|
TABLE |
| Processing_log |
• |
• |
|
|
|
TABLE |
| ProcessingMaterialCategory |
User |
• |
• |
|
|
TABLE |
| ProjectAnalysis |
User |
• |
|
|
|
TABLE |
| ProjectMaterialCategory |
User |
|
|
|
|
TABLE |
| ProjectProcessing |
User |
• |
|
|
|
TABLE |
| ProjectTaxonomicGroup |
User |
|
|
|
|
TABLE |
| ProjectUser |
User |
|
|
|
|
TABLE |
| Property |
User |
|
|
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
User |
|
|
|
|
TABLE |
| ReplicationPublisher |
• |
|
|
|
|
TABLE |
| Task |
User |
|
|
|
|
TABLE |
| Task_log |
|
|
|
|
|
TABLE |
| TaskDateType_Enum |
User |
|
|
|
|
TABLE |
| TaskModule |
User |
|
|
|
|
TABLE |
| TaskModule_log |
|
|
|
|
|
TABLE |
| TaskModuleType_Enum |
User |
|
|
|
|
TABLE |
| TaskResult |
User |
|
|
|
|
TABLE |
| TaskResult_log |
|
|
|
|
|
TABLE |
| TaskType_Enum |
User |
|
|
|
|
TABLE |
| Transaction |
User |
|
|
|
|
TABLE |
| Transaction_log |
Typist |
Typist |
|
|
|
TABLE |
| TransactionAgent |
|
|
|
|
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
User |
|
|
|
|
TABLE |
| TransactionDocument |
User |
Typist |
Typist |
|
|
TABLE |
| TransactionDocument_log |
Typist |
Typist |
|
|
|
TABLE |
| TransactionPayment |
|
|
|
|
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
User |
|
|
|
|
VIEW |
| AnnotationPart |
User |
|
|
|
|
VIEW |
| AnnotationSpecimen |
User |
|
|
|
|
VIEW |
| AnnotationUnit |
User |
|
|
|
|
VIEW |
| CollectionAgent_Core |
User |
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
User |
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
User |
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
User |
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
|
|
|
|
|
VIEW |
| FirstLinesIdentification |
User |
|
|
|
|
VIEW |
| Identification_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
User |
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
User |
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
User |
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
User |
|
|
|
|
VIEW |
| TransactionList_H7 |
User |
|
|
|
|
VIEW |
| TransactionPermit |
User |
|
|
|
|
VIEW |
| TransactionRegulation |
User |
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
User |
|
|
|
|
VIEW |
| ViewCollectionEventImage |
User |
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
User |
|
|
|
|
VIEW |
| ViewCollectionImage |
User |
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
User |
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
User |
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
User |
|
|
|
|
VIEW |
| AgentOneString |
• |
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
User |
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
User |
|
|
|
|
FUNCTION |
| AnalysisList |
User |
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
User |
|
|
|
|
FUNCTION |
| AnalysisProjectList |
User |
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
User |
FUNCTION |
| CollCharacterType_List |
• |
|
|
|
|
FUNCTION |
| CollDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollectionChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
User |
|
|
|
|
FUNCTION |
| CollectionLocation |
User |
|
|
|
|
FUNCTION |
| CollectionLocationAll |
User |
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
User |
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
User |
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
User |
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
• |
|
|
|
|
FUNCTION |
| CollEventImageType_List |
• |
|
|
|
|
FUNCTION |
| CollExchangeType_List |
• |
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
• |
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
User |
|
|
|
|
FUNCTION |
| CollLabelType_List |
User |
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
User |
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
User |
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
User |
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
User |
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
• |
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
|
|
|
|
|
FUNCTION |
| CurrentUser |
User |
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
User |
FUNCTION |
| DefaultProjectID |
|
|
|
|
User |
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
User |
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
User |
FUNCTION |
| EntityInformation_2 |
User |
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
User |
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
User |
FUNCTION |
| EventSeriesChildNodes |
User |
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
User |
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
User |
FUNCTION |
| EventSuperiorList |
User |
|
|
|
|
FUNCTION |
| FirstLines_4 |
User |
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesSeries |
User |
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
User |
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
User |
|
|
|
|
FUNCTION |
| ManagerCollectionList |
|
|
|
|
|
FUNCTION |
| MethodChildNodes |
User |
|
|
|
|
FUNCTION |
| MethodHierarchy |
User |
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
User |
|
|
|
|
FUNCTION |
| NameList |
User |
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
User |
|
|
|
|
FUNCTION |
| NameListPlants |
User |
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
Typist |
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
• |
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
User |
FUNCTION |
| ProcessingChildNodes |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
User |
|
|
|
|
FUNCTION |
| ProcessingListForPart |
User |
|
|
|
|
FUNCTION |
| ProcessingProjectList |
User |
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
User |
FUNCTION |
| RequesterCollectionList |
|
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
User |
FUNCTION |
| TaskChildNodes |
User |
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaskHierarchy |
User |
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
User |
FUNCTION |
| TransactionChildNodes |
User |
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
User |
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
User |
FUNCTION |
| TransactionHierarchy |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| UserCollectionList |
User |
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
User |
FUNCTION |
| Version |
|
|
|
|
User |
FUNCTION |
| VersionClient |
|
|
|
|
User |
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
User |
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
• |
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
• |
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
User |
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
• |
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
• |
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| SetXmlValue |
|
|
|
|
User |
PROCEDURE |
| Inheriting from roles: |
|
|
|
|
|
|
Role RegulationManager
Full access for tables CollectionEventRegulation and Regulation
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
|
|
|
|
|
TABLE |
| Analysis_log |
|
|
|
|
|
TABLE |
| AnalysisResult |
|
|
|
|
|
TABLE |
| AnalysisTaxonomicGroup |
|
|
|
|
|
TABLE |
| Annotation |
|
|
|
|
|
TABLE |
| Annotation_log |
|
|
|
|
|
TABLE |
| AnnotationType_Enum |
|
|
|
|
|
TABLE |
| AnonymCollector |
|
|
|
|
|
TABLE |
| CacheDatabase2 |
|
|
|
|
|
TABLE |
| CacheDescription |
|
|
|
|
|
TABLE |
| CollCircumstances_Enum |
|
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
|
|
|
|
|
TABLE |
| CollCollectionType_Enum |
|
|
|
|
|
TABLE |
| CollDateCategory_Enum |
|
|
|
|
|
TABLE |
| Collection |
|
|
|
|
|
TABLE |
| Collection_log |
|
|
|
|
|
TABLE |
| CollectionAgent |
|
|
|
|
|
TABLE |
| CollectionAgent_log |
|
|
|
|
|
TABLE |
| CollectionEvent |
|
|
|
|
|
TABLE |
| CollectionEvent_log |
|
|
|
|
|
TABLE |
| CollectionEventImage |
|
|
|
|
|
TABLE |
| CollectionEventImage_log |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation_log |
|
|
|
|
|
TABLE |
| CollectionEventMethod |
|
|
|
|
|
TABLE |
| CollectionEventMethod_log |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue_log |
|
|
|
|
|
TABLE |
| CollectionEventProperty |
|
|
|
|
|
TABLE |
| CollectionEventProperty_log |
|
|
|
|
|
TABLE |
| CollectionEventRegulation |
|
• |
• |
• |
|
TABLE |
| CollectionEventRegulation_log |
|
|
|
|
|
TABLE |
| CollectionEventSeries |
|
|
|
|
|
TABLE |
| CollectionEventSeries_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
|
|
|
|
|
TABLE |
| CollectionImage_log |
|
|
|
|
|
TABLE |
| CollectionManager |
|
|
|
|
|
TABLE |
| CollectionProject |
|
|
|
|
|
TABLE |
| CollectionProject_log |
|
|
|
|
|
TABLE |
| CollectionSpecimen |
|
|
|
|
|
TABLE |
| CollectionSpecimen_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction_log |
|
|
|
|
|
TABLE |
| CollectionTask |
|
|
|
|
|
TABLE |
| CollectionTask_log |
|
|
|
|
|
TABLE |
| CollectionTaskImage |
|
|
|
|
|
TABLE |
| CollectionTaskImage_log |
|
|
|
|
|
TABLE |
| CollectionTaskMetric |
|
|
|
|
|
TABLE |
| CollectionTaskMetric_log |
|
|
|
|
|
TABLE |
| CollEventDateCategory_Enum |
|
|
|
|
|
TABLE |
| CollEventImageType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
|
|
|
|
|
TABLE |
| CollExchangeType_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationCategory_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationDateCategory_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationQualifier_Enum |
|
|
|
|
|
TABLE |
| CollLabelTranscriptionState_Enum |
|
|
|
|
|
TABLE |
| CollLabelType_Enum |
|
|
|
|
|
TABLE |
| CollMaterialCategory_Enum |
|
|
|
|
|
TABLE |
| CollRetrievalType_Enum |
|
|
|
|
|
TABLE |
| CollSpecimenImageType_Enum |
|
|
|
|
|
TABLE |
| CollSpecimenRelationType_Enum |
|
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
|
|
|
|
|
TABLE |
| CollTaxonomicGroup_Enum |
|
|
|
|
|
TABLE |
| CollTransactionType_Enum |
|
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
|
|
|
|
|
TABLE |
| CollUnitRelationType_Enum |
|
|
|
|
|
TABLE |
| Entity |
|
|
|
|
|
TABLE |
| EntityAccessibility_Enum |
|
|
|
|
|
TABLE |
| EntityContext_Enum |
|
|
|
|
|
TABLE |
| EntityDetermination_Enum |
|
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
|
|
|
|
|
TABLE |
| EntityRepresentation |
|
|
|
|
|
TABLE |
| EntityUsage |
|
|
|
|
|
TABLE |
| EntityUsage_Enum |
|
|
|
|
|
TABLE |
| EntityVisibility_Enum |
|
|
|
|
|
TABLE |
| ExternalIdentifier |
|
|
|
|
|
TABLE |
| ExternalIdentifier_log |
|
|
|
|
|
TABLE |
| ExternalIdentifierType |
|
|
|
|
|
TABLE |
| ExternalIdentifierType_log |
|
|
|
|
|
TABLE |
| Identification |
|
|
|
|
|
TABLE |
| Identification_log |
|
|
|
|
|
TABLE |
| IdentificationUnit |
|
|
|
|
|
TABLE |
| IdentificationUnit_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart_log |
|
|
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
|
|
|
|
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
|
|
|
|
|
TABLE |
| Method_log |
|
|
|
|
|
TABLE |
| MethodForAnalysis |
|
|
|
|
|
TABLE |
| MethodForProcessing |
|
|
|
|
|
TABLE |
| Parameter |
|
|
|
|
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
|
|
|
|
|
TABLE |
| Processing |
|
|
|
|
|
TABLE |
| Processing_log |
|
|
|
|
|
TABLE |
| ProcessingMaterialCategory |
|
|
|
|
|
TABLE |
| ProjectAnalysis |
|
|
|
|
|
TABLE |
| ProjectMaterialCategory |
|
|
|
|
|
TABLE |
| ProjectProcessing |
|
|
|
|
|
TABLE |
| ProjectTaxonomicGroup |
|
|
|
|
|
TABLE |
| ProjectUser |
|
|
|
|
|
TABLE |
| Property |
|
|
|
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
|
|
|
|
|
TABLE |
| ReplicationPublisher |
|
|
|
|
|
TABLE |
| Task |
|
|
|
|
|
TABLE |
| Task_log |
|
|
|
|
|
TABLE |
| TaskDateType_Enum |
|
|
|
|
|
TABLE |
| TaskModule |
|
|
|
|
|
TABLE |
| TaskModule_log |
|
|
|
|
|
TABLE |
| TaskModuleType_Enum |
|
|
|
|
|
TABLE |
| TaskResult |
|
|
|
|
|
TABLE |
| TaskResult_log |
|
|
|
|
|
TABLE |
| TaskType_Enum |
|
|
|
|
|
TABLE |
| Transaction |
|
|
|
|
|
TABLE |
| Transaction_log |
|
|
|
|
|
TABLE |
| TransactionAgent |
|
|
|
|
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
|
|
|
|
|
TABLE |
| TransactionDocument |
|
|
|
|
|
TABLE |
| TransactionDocument_log |
|
|
|
|
|
TABLE |
| TransactionPayment |
|
|
|
|
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
|
|
|
|
|
VIEW |
| AnnotationPart |
|
|
|
|
|
VIEW |
| AnnotationSpecimen |
|
|
|
|
|
VIEW |
| AnnotationUnit |
|
|
|
|
|
VIEW |
| CollectionAgent_Core |
|
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
|
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
|
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
|
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
|
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
|
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
|
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
|
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
|
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
|
|
|
|
|
VIEW |
| FirstLinesIdentification |
|
|
|
|
|
VIEW |
| Identification_Core2 |
|
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
|
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
|
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
|
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
|
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
|
|
|
|
|
VIEW |
| TransactionList_H7 |
|
|
|
|
|
VIEW |
| TransactionPermit |
|
|
|
|
|
VIEW |
| TransactionRegulation |
|
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
|
|
|
|
|
VIEW |
| ViewCollectionEventImage |
|
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
|
|
|
|
|
VIEW |
| ViewCollectionImage |
|
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
|
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
|
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
|
|
|
|
|
VIEW |
| AgentOneString |
|
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
|
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
|
|
|
|
|
FUNCTION |
| AnalysisList |
|
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
|
|
|
|
|
FUNCTION |
| AnalysisProjectList |
|
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
|
FUNCTION |
| CollCharacterType_List |
|
|
|
|
|
FUNCTION |
| CollDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollectionChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
|
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
|
|
|
|
|
FUNCTION |
| CollectionLocation |
|
|
|
|
|
FUNCTION |
| CollectionLocationAll |
|
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
|
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
|
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
|
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollEventImageType_List |
|
|
|
|
|
FUNCTION |
| CollExchangeType_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
|
|
|
|
|
FUNCTION |
| CollLabelType_List |
|
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
|
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
|
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
|
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
|
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
|
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
|
|
|
|
|
FUNCTION |
| CurrentUser |
|
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
|
FUNCTION |
| DefaultProjectID |
|
|
|
|
|
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
|
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
|
FUNCTION |
| EntityInformation_2 |
|
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
|
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
|
FUNCTION |
| EventSeriesChildNodes |
|
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
|
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
|
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
|
FUNCTION |
| EventSuperiorList |
|
|
|
|
|
FUNCTION |
| FirstLines_4 |
|
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
|
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
|
|
|
|
|
FUNCTION |
| FirstLinesSeries |
|
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
|
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
|
|
|
|
|
FUNCTION |
| ManagerCollectionList |
|
|
|
|
|
FUNCTION |
| MethodChildNodes |
|
|
|
|
|
FUNCTION |
| MethodHierarchy |
|
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
|
|
|
|
|
FUNCTION |
| NameList |
|
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
|
|
|
|
|
FUNCTION |
| NameListPlants |
|
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
|
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
|
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
|
FUNCTION |
| ProcessingChildNodes |
|
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
|
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
|
|
|
|
|
FUNCTION |
| ProcessingListForPart |
|
|
|
|
|
FUNCTION |
| ProcessingProjectList |
|
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
|
FUNCTION |
| RequesterCollectionList |
|
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
|
FUNCTION |
| TaskChildNodes |
|
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
|
FUNCTION |
| TaskHierarchy |
|
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
|
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
|
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
|
FUNCTION |
| TransactionChildNodes |
|
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
|
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
|
FUNCTION |
| TransactionHierarchy |
|
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
|
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
|
|
|
|
|
FUNCTION |
| UserCollectionList |
|
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
|
FUNCTION |
| Version |
|
|
|
|
|
FUNCTION |
| VersionClient |
|
|
|
|
|
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
|
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
|
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
|
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
|
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
|
PROCEDURE |
| SetXmlValue |
|
|
|
|
|
PROCEDURE |
Role Replicator
Role with special rights needed for replication
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
|
|
|
|
|
TABLE |
| Analysis_log |
|
|
|
|
|
TABLE |
| AnalysisResult |
|
|
|
|
|
TABLE |
| AnalysisTaxonomicGroup |
|
|
|
|
|
TABLE |
| Annotation |
|
|
|
|
|
TABLE |
| Annotation_log |
|
|
|
|
|
TABLE |
| AnnotationType_Enum |
|
|
|
|
|
TABLE |
| AnonymCollector |
|
|
|
|
|
TABLE |
| CacheDatabase2 |
|
|
|
|
|
TABLE |
| CacheDescription |
|
|
|
|
|
TABLE |
| CollCircumstances_Enum |
|
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
|
|
|
|
|
TABLE |
| CollCollectionType_Enum |
|
|
|
|
|
TABLE |
| CollDateCategory_Enum |
|
|
|
|
|
TABLE |
| Collection |
|
|
|
|
|
TABLE |
| Collection_log |
|
|
|
|
|
TABLE |
| CollectionAgent |
|
|
|
|
|
TABLE |
| CollectionAgent_log |
|
|
|
|
|
TABLE |
| CollectionEvent |
|
|
|
|
|
TABLE |
| CollectionEvent_log |
|
|
|
|
|
TABLE |
| CollectionEventImage |
|
|
|
|
|
TABLE |
| CollectionEventImage_log |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation_log |
|
|
|
|
|
TABLE |
| CollectionEventMethod |
|
|
|
|
|
TABLE |
| CollectionEventMethod_log |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue_log |
|
|
|
|
|
TABLE |
| CollectionEventProperty |
|
|
|
|
|
TABLE |
| CollectionEventProperty_log |
|
|
|
|
|
TABLE |
| CollectionEventRegulation |
|
|
|
|
|
TABLE |
| CollectionEventRegulation_log |
|
|
|
|
|
TABLE |
| CollectionEventSeries |
|
|
|
|
|
TABLE |
| CollectionEventSeries_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
|
|
|
|
|
TABLE |
| CollectionImage_log |
|
|
|
|
|
TABLE |
| CollectionManager |
|
|
|
|
|
TABLE |
| CollectionProject |
|
|
|
|
|
TABLE |
| CollectionProject_log |
|
|
|
|
|
TABLE |
| CollectionSpecimen |
|
|
|
|
|
TABLE |
| CollectionSpecimen_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction_log |
|
|
|
|
|
TABLE |
| CollectionTask |
|
|
|
|
|
TABLE |
| CollectionTask_log |
|
|
|
|
|
TABLE |
| CollectionTaskImage |
|
|
|
|
|
TABLE |
| CollectionTaskImage_log |
|
|
|
|
|
TABLE |
| CollectionTaskMetric |
|
|
|
|
|
TABLE |
| CollectionTaskMetric_log |
|
|
|
|
|
TABLE |
| CollEventDateCategory_Enum |
|
|
|
|
|
TABLE |
| CollEventImageType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
|
|
|
|
|
TABLE |
| CollExchangeType_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationCategory_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationDateCategory_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationQualifier_Enum |
|
|
|
|
|
TABLE |
| CollLabelTranscriptionState_Enum |
|
|
|
|
|
TABLE |
| CollLabelType_Enum |
|
|
|
|
|
TABLE |
| CollMaterialCategory_Enum |
|
|
|
|
|
TABLE |
| CollRetrievalType_Enum |
|
|
|
|
|
TABLE |
| CollSpecimenImageType_Enum |
|
|
|
|
|
TABLE |
| CollSpecimenRelationType_Enum |
|
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
|
|
|
|
|
TABLE |
| CollTaxonomicGroup_Enum |
|
|
|
|
|
TABLE |
| CollTransactionType_Enum |
|
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
|
|
|
|
|
TABLE |
| CollUnitRelationType_Enum |
|
|
|
|
|
TABLE |
| Entity |
|
|
|
|
|
TABLE |
| EntityAccessibility_Enum |
|
|
|
|
|
TABLE |
| EntityContext_Enum |
|
|
|
|
|
TABLE |
| EntityDetermination_Enum |
|
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
|
|
|
|
|
TABLE |
| EntityRepresentation |
|
|
|
|
|
TABLE |
| EntityUsage |
|
|
|
|
|
TABLE |
| EntityUsage_Enum |
|
|
|
|
|
TABLE |
| EntityVisibility_Enum |
|
|
|
|
|
TABLE |
| ExternalIdentifier |
|
|
|
|
|
TABLE |
| ExternalIdentifier_log |
|
|
|
|
|
TABLE |
| ExternalIdentifierType |
|
|
|
|
|
TABLE |
| ExternalIdentifierType_log |
|
|
|
|
|
TABLE |
| Identification |
|
|
|
|
|
TABLE |
| Identification_log |
|
|
|
|
|
TABLE |
| IdentificationUnit |
|
|
|
|
|
TABLE |
| IdentificationUnit_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart_log |
|
|
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
|
|
|
|
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
|
|
|
|
|
TABLE |
| Method_log |
|
|
|
|
|
TABLE |
| MethodForAnalysis |
|
|
|
|
|
TABLE |
| MethodForProcessing |
|
|
|
|
|
TABLE |
| Parameter |
|
|
|
|
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
|
|
|
|
|
TABLE |
| Processing |
|
|
|
|
|
TABLE |
| Processing_log |
|
|
|
|
|
TABLE |
| ProcessingMaterialCategory |
|
|
|
|
|
TABLE |
| ProjectAnalysis |
|
|
|
|
|
TABLE |
| ProjectMaterialCategory |
|
|
|
|
|
TABLE |
| ProjectProcessing |
|
|
|
|
|
TABLE |
| ProjectTaxonomicGroup |
|
|
|
|
|
TABLE |
| ProjectUser |
|
|
|
|
|
TABLE |
| Property |
|
|
|
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
|
|
|
|
|
TABLE |
| ReplicationPublisher |
|
|
|
|
|
TABLE |
| Task |
|
|
|
|
|
TABLE |
| Task_log |
|
|
|
|
|
TABLE |
| TaskDateType_Enum |
|
|
|
|
|
TABLE |
| TaskModule |
|
|
|
|
|
TABLE |
| TaskModule_log |
|
|
|
|
|
TABLE |
| TaskModuleType_Enum |
|
|
|
|
|
TABLE |
| TaskResult |
|
|
|
|
|
TABLE |
| TaskResult_log |
|
|
|
|
|
TABLE |
| TaskType_Enum |
|
|
|
|
|
TABLE |
| Transaction |
|
|
|
|
|
TABLE |
| Transaction_log |
|
|
|
|
|
TABLE |
| TransactionAgent |
|
|
|
|
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
|
|
|
|
|
TABLE |
| TransactionDocument |
|
|
|
|
|
TABLE |
| TransactionDocument_log |
|
|
|
|
|
TABLE |
| TransactionPayment |
|
|
|
|
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
|
|
|
|
|
VIEW |
| AnnotationPart |
|
|
|
|
|
VIEW |
| AnnotationSpecimen |
|
|
|
|
|
VIEW |
| AnnotationUnit |
|
|
|
|
|
VIEW |
| CollectionAgent_Core |
|
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
|
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
|
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
|
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
|
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
|
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
|
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
|
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
|
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
|
|
|
|
|
VIEW |
| FirstLinesIdentification |
|
|
|
|
|
VIEW |
| Identification_Core2 |
|
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
|
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
|
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
|
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
|
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
|
|
|
|
|
VIEW |
| TransactionList_H7 |
|
|
|
|
|
VIEW |
| TransactionPermit |
|
|
|
|
|
VIEW |
| TransactionRegulation |
|
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
|
|
|
|
|
VIEW |
| ViewCollectionEventImage |
|
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
|
|
|
|
|
VIEW |
| ViewCollectionImage |
|
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
|
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
|
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
|
|
|
|
|
VIEW |
| AgentOneString |
|
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
|
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
|
|
|
|
|
FUNCTION |
| AnalysisList |
|
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
|
|
|
|
|
FUNCTION |
| AnalysisProjectList |
|
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
|
FUNCTION |
| CollCharacterType_List |
|
|
|
|
|
FUNCTION |
| CollDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollectionChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
|
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
|
|
|
|
|
FUNCTION |
| CollectionLocation |
|
|
|
|
|
FUNCTION |
| CollectionLocationAll |
|
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
|
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
|
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
|
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollEventImageType_List |
|
|
|
|
|
FUNCTION |
| CollExchangeType_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
|
|
|
|
|
FUNCTION |
| CollLabelType_List |
|
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
|
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
|
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
|
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
|
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
|
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
|
|
|
|
|
FUNCTION |
| CurrentUser |
|
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
|
FUNCTION |
| DefaultProjectID |
|
|
|
|
|
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
|
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
|
FUNCTION |
| EntityInformation_2 |
|
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
|
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
|
FUNCTION |
| EventSeriesChildNodes |
|
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
|
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
|
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
|
FUNCTION |
| EventSuperiorList |
|
|
|
|
|
FUNCTION |
| FirstLines_4 |
|
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
|
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
|
|
|
|
|
FUNCTION |
| FirstLinesSeries |
|
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
|
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
|
|
|
|
|
FUNCTION |
| ManagerCollectionList |
|
|
|
|
|
FUNCTION |
| MethodChildNodes |
|
|
|
|
|
FUNCTION |
| MethodHierarchy |
|
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
|
|
|
|
|
FUNCTION |
| NameList |
|
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
|
|
|
|
|
FUNCTION |
| NameListPlants |
|
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
|
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
|
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
|
FUNCTION |
| ProcessingChildNodes |
|
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
|
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
|
|
|
|
|
FUNCTION |
| ProcessingListForPart |
|
|
|
|
|
FUNCTION |
| ProcessingProjectList |
|
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
|
FUNCTION |
| RequesterCollectionList |
|
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
|
FUNCTION |
| TaskChildNodes |
|
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
|
FUNCTION |
| TaskHierarchy |
|
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
|
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
|
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
|
FUNCTION |
| TransactionChildNodes |
|
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
|
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
|
FUNCTION |
| TransactionHierarchy |
|
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
|
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
|
|
|
|
|
FUNCTION |
| UserCollectionList |
|
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
|
FUNCTION |
| Version |
|
|
|
|
|
FUNCTION |
| VersionClient |
|
|
|
|
|
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
|
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
|
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
|
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
|
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
|
PROCEDURE |
| SetXmlValue |
|
|
|
|
|
PROCEDURE |
Role Requester
Role with the permission to place requests for loans
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
|
|
|
|
|
TABLE |
| Analysis_log |
|
|
|
|
|
TABLE |
| AnalysisResult |
|
|
|
|
|
TABLE |
| AnalysisTaxonomicGroup |
|
|
|
|
|
TABLE |
| Annotation |
|
|
|
|
|
TABLE |
| Annotation_log |
|
|
|
|
|
TABLE |
| AnnotationType_Enum |
|
|
|
|
|
TABLE |
| AnonymCollector |
|
|
|
|
|
TABLE |
| CacheDatabase2 |
|
|
|
|
|
TABLE |
| CacheDescription |
|
|
|
|
|
TABLE |
| CollCircumstances_Enum |
|
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
|
|
|
|
|
TABLE |
| CollCollectionType_Enum |
|
|
|
|
|
TABLE |
| CollDateCategory_Enum |
|
|
|
|
|
TABLE |
| Collection |
|
|
|
|
|
TABLE |
| Collection_log |
|
|
|
|
|
TABLE |
| CollectionAgent |
|
|
|
|
|
TABLE |
| CollectionAgent_log |
|
|
|
|
|
TABLE |
| CollectionEvent |
|
|
|
|
|
TABLE |
| CollectionEvent_log |
|
|
|
|
|
TABLE |
| CollectionEventImage |
|
|
|
|
|
TABLE |
| CollectionEventImage_log |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation_log |
|
|
|
|
|
TABLE |
| CollectionEventMethod |
|
|
|
|
|
TABLE |
| CollectionEventMethod_log |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue_log |
|
|
|
|
|
TABLE |
| CollectionEventProperty |
|
|
|
|
|
TABLE |
| CollectionEventProperty_log |
|
|
|
|
|
TABLE |
| CollectionEventRegulation |
|
|
|
|
|
TABLE |
| CollectionEventRegulation_log |
|
|
|
|
|
TABLE |
| CollectionEventSeries |
|
|
|
|
|
TABLE |
| CollectionEventSeries_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
|
|
|
|
|
TABLE |
| CollectionImage_log |
|
|
|
|
|
TABLE |
| CollectionManager |
• |
|
|
|
|
TABLE |
| CollectionProject |
|
|
|
|
|
TABLE |
| CollectionProject_log |
|
|
|
|
|
TABLE |
| CollectionSpecimen |
|
|
|
|
|
TABLE |
| CollectionSpecimen_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction_log |
|
|
|
|
|
TABLE |
| CollectionTask |
|
|
|
|
|
TABLE |
| CollectionTask_log |
|
|
|
|
|
TABLE |
| CollectionTaskImage |
|
|
|
|
|
TABLE |
| CollectionTaskImage_log |
|
|
|
|
|
TABLE |
| CollectionTaskMetric |
|
|
|
|
|
TABLE |
| CollectionTaskMetric_log |
|
|
|
|
|
TABLE |
| CollEventDateCategory_Enum |
|
|
|
|
|
TABLE |
| CollEventImageType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
|
|
|
|
|
TABLE |
| CollExchangeType_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationCategory_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationDateCategory_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationQualifier_Enum |
|
|
|
|
|
TABLE |
| CollLabelTranscriptionState_Enum |
|
|
|
|
|
TABLE |
| CollLabelType_Enum |
|
|
|
|
|
TABLE |
| CollMaterialCategory_Enum |
|
|
|
|
|
TABLE |
| CollRetrievalType_Enum |
|
|
|
|
|
TABLE |
| CollSpecimenImageType_Enum |
|
|
|
|
|
TABLE |
| CollSpecimenRelationType_Enum |
|
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
|
|
|
|
|
TABLE |
| CollTaxonomicGroup_Enum |
|
|
|
|
|
TABLE |
| CollTransactionType_Enum |
|
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
|
|
|
|
|
TABLE |
| CollUnitRelationType_Enum |
|
|
|
|
|
TABLE |
| Entity |
|
|
|
|
|
TABLE |
| EntityAccessibility_Enum |
|
|
|
|
|
TABLE |
| EntityContext_Enum |
|
|
|
|
|
TABLE |
| EntityDetermination_Enum |
|
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
|
|
|
|
|
TABLE |
| EntityRepresentation |
|
|
|
|
|
TABLE |
| EntityUsage |
|
|
|
|
|
TABLE |
| EntityUsage_Enum |
|
|
|
|
|
TABLE |
| EntityVisibility_Enum |
|
|
|
|
|
TABLE |
| ExternalIdentifier |
|
|
|
|
|
TABLE |
| ExternalIdentifier_log |
|
|
|
|
|
TABLE |
| ExternalIdentifierType |
|
|
|
|
|
TABLE |
| ExternalIdentifierType_log |
|
|
|
|
|
TABLE |
| Identification |
|
|
|
|
|
TABLE |
| Identification_log |
|
|
|
|
|
TABLE |
| IdentificationUnit |
|
|
|
|
|
TABLE |
| IdentificationUnit_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart_log |
|
|
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
|
|
|
|
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
|
|
|
|
|
TABLE |
| Method_log |
|
|
|
|
|
TABLE |
| MethodForAnalysis |
|
|
|
|
|
TABLE |
| MethodForProcessing |
|
|
|
|
|
TABLE |
| Parameter |
|
|
|
|
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
|
|
|
|
|
TABLE |
| Processing |
|
|
|
|
|
TABLE |
| Processing_log |
|
|
|
|
|
TABLE |
| ProcessingMaterialCategory |
|
|
|
|
|
TABLE |
| ProjectAnalysis |
|
|
|
|
|
TABLE |
| ProjectMaterialCategory |
|
|
|
|
|
TABLE |
| ProjectProcessing |
|
|
|
|
|
TABLE |
| ProjectTaxonomicGroup |
|
|
|
|
|
TABLE |
| ProjectUser |
|
|
|
|
|
TABLE |
| Property |
|
|
|
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
|
|
|
|
|
TABLE |
| ReplicationPublisher |
|
|
|
|
|
TABLE |
| Task |
|
|
|
|
|
TABLE |
| Task_log |
|
|
|
|
|
TABLE |
| TaskDateType_Enum |
|
|
|
|
|
TABLE |
| TaskModule |
|
|
|
|
|
TABLE |
| TaskModule_log |
|
|
|
|
|
TABLE |
| TaskModuleType_Enum |
|
|
|
|
|
TABLE |
| TaskResult |
|
|
|
|
|
TABLE |
| TaskResult_log |
|
|
|
|
|
TABLE |
| TaskType_Enum |
|
|
|
|
|
TABLE |
| Transaction |
|
|
|
|
|
TABLE |
| Transaction_log |
|
|
|
|
|
TABLE |
| TransactionAgent |
|
|
|
|
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
|
|
|
|
|
TABLE |
| TransactionDocument |
|
|
|
|
|
TABLE |
| TransactionDocument_log |
|
|
|
|
|
TABLE |
| TransactionPayment |
|
|
|
|
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
|
|
|
|
|
VIEW |
| AnnotationPart |
|
|
|
|
|
VIEW |
| AnnotationSpecimen |
|
|
|
|
|
VIEW |
| AnnotationUnit |
|
|
|
|
|
VIEW |
| CollectionAgent_Core |
|
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
|
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
|
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
|
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
|
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
|
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
|
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
|
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
|
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
• |
|
|
|
|
VIEW |
| FirstLinesIdentification |
|
|
|
|
|
VIEW |
| Identification_Core2 |
|
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
|
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
|
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
|
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
|
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
|
|
|
|
|
VIEW |
| TransactionList_H7 |
|
|
|
|
|
VIEW |
| TransactionPermit |
|
|
|
|
|
VIEW |
| TransactionRegulation |
|
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
|
|
|
|
|
VIEW |
| ViewCollectionEventImage |
|
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
|
|
|
|
|
VIEW |
| ViewCollectionImage |
|
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
|
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
|
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
|
|
|
|
|
VIEW |
| AgentOneString |
|
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
|
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
|
|
|
|
|
FUNCTION |
| AnalysisList |
|
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
|
|
|
|
|
FUNCTION |
| AnalysisProjectList |
|
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
|
FUNCTION |
| CollCharacterType_List |
|
|
|
|
|
FUNCTION |
| CollDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollectionChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
|
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
|
|
|
|
|
FUNCTION |
| CollectionLocation |
|
|
|
|
|
FUNCTION |
| CollectionLocationAll |
|
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
|
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
|
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
|
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollEventImageType_List |
|
|
|
|
|
FUNCTION |
| CollExchangeType_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
|
|
|
|
|
FUNCTION |
| CollLabelType_List |
|
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
|
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
|
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
|
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
|
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
|
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
|
|
|
|
|
FUNCTION |
| CurrentUser |
|
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
|
FUNCTION |
| DefaultProjectID |
|
|
|
|
|
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
|
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
|
FUNCTION |
| EntityInformation_2 |
|
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
|
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
|
FUNCTION |
| EventSeriesChildNodes |
|
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
|
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
|
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
|
FUNCTION |
| EventSuperiorList |
|
|
|
|
|
FUNCTION |
| FirstLines_4 |
|
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
|
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
|
|
|
|
|
FUNCTION |
| FirstLinesSeries |
|
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
|
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
|
|
|
|
|
FUNCTION |
| ManagerCollectionList |
|
|
|
|
|
FUNCTION |
| MethodChildNodes |
|
|
|
|
|
FUNCTION |
| MethodHierarchy |
|
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
|
|
|
|
|
FUNCTION |
| NameList |
|
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
|
|
|
|
|
FUNCTION |
| NameListPlants |
|
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
|
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
|
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
|
FUNCTION |
| ProcessingChildNodes |
|
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
|
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
|
|
|
|
|
FUNCTION |
| ProcessingListForPart |
|
|
|
|
|
FUNCTION |
| ProcessingProjectList |
|
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
|
FUNCTION |
| RequesterCollectionList |
• |
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
|
FUNCTION |
| TaskChildNodes |
|
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
|
FUNCTION |
| TaskHierarchy |
|
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
|
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
|
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
|
FUNCTION |
| TransactionChildNodes |
|
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
|
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
|
FUNCTION |
| TransactionHierarchy |
|
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
|
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
|
|
|
|
|
FUNCTION |
| UserCollectionList |
|
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
|
FUNCTION |
| Version |
|
|
|
|
|
FUNCTION |
| VersionClient |
|
|
|
|
|
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
|
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
|
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
|
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
|
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
|
PROCEDURE |
| SetXmlValue |
|
|
|
|
|
PROCEDURE |
Role StorageManager
Role for handling Reference References and related information
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
User |
|
|
|
|
TABLE |
| Analysis_log |
|
|
|
|
|
TABLE |
| AnalysisResult |
User |
|
|
|
|
TABLE |
| AnalysisTaxonomicGroup |
User |
|
|
|
|
TABLE |
| Annotation |
User |
User |
User |
|
|
TABLE |
| Annotation_log |
|
|
|
|
|
TABLE |
| AnnotationType_Enum |
User |
|
|
|
|
TABLE |
| AnonymCollector |
|
|
|
|
|
TABLE |
| CacheDatabase2 |
|
|
|
|
|
TABLE |
| CacheDescription |
User |
User |
User |
User |
|
TABLE |
| CollCircumstances_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionType_Enum |
User |
|
|
|
|
TABLE |
| CollDateCategory_Enum |
User |
|
|
|
|
TABLE |
| Collection |
User |
|
|
|
|
TABLE |
| Collection_log |
|
|
|
|
|
TABLE |
| CollectionAgent |
User |
|
|
|
|
TABLE |
| CollectionAgent_log |
|
|
|
|
|
TABLE |
| CollectionEvent |
User |
|
|
|
|
TABLE |
| CollectionEvent_log |
|
|
|
|
|
TABLE |
| CollectionEventImage |
User |
|
|
|
|
TABLE |
| CollectionEventImage_log |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation |
User |
|
|
|
|
TABLE |
| CollectionEventLocalisation_log |
|
|
|
|
|
TABLE |
| CollectionEventMethod |
User |
|
|
|
|
TABLE |
| CollectionEventMethod_log |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue |
User |
|
|
|
|
TABLE |
| CollectionEventParameterValue_log |
|
|
|
|
|
TABLE |
| CollectionEventProperty |
User |
|
|
|
|
TABLE |
| CollectionEventProperty_log |
|
|
|
|
|
TABLE |
| CollectionEventRegulation |
User |
|
|
|
|
TABLE |
| CollectionEventRegulation_log |
|
|
|
|
|
TABLE |
| CollectionEventSeries |
User |
|
|
|
|
TABLE |
| CollectionEventSeries_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
User |
|
|
|
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
User |
|
|
|
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
User |
|
|
|
|
TABLE |
| CollectionImage_log |
|
|
|
|
|
TABLE |
| CollectionManager |
|
|
|
|
|
TABLE |
| CollectionProject |
User |
|
|
|
|
TABLE |
| CollectionProject_log |
|
|
|
|
|
TABLE |
| CollectionSpecimen |
User |
|
|
|
|
TABLE |
| CollectionSpecimen_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage |
User |
|
|
|
|
TABLE |
| CollectionSpecimenImage_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
User |
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart |
User |
• |
• |
• |
|
TABLE |
| CollectionSpecimenPart_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
User |
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing |
User |
• |
• |
• |
|
TABLE |
| CollectionSpecimenProcessing_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
User |
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
User |
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
User |
|
|
|
|
TABLE |
| CollectionSpecimenReference_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation |
User |
|
|
|
|
TABLE |
| CollectionSpecimenRelation_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction |
User |
• |
• |
|
|
TABLE |
| CollectionSpecimenTransaction_log |
|
|
|
|
|
TABLE |
| CollectionTask |
User |
|
|
|
|
TABLE |
| CollectionTask_log |
|
|
|
|
|
TABLE |
| CollectionTaskImage |
User |
|
|
|
|
TABLE |
| CollectionTaskImage_log |
|
|
|
|
|
TABLE |
| CollectionTaskMetric |
User |
|
|
|
|
TABLE |
| CollectionTaskMetric_log |
|
|
|
|
|
TABLE |
| CollEventDateCategory_Enum |
User |
|
|
|
|
TABLE |
| CollEventImageType_Enum |
User |
|
|
|
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
User |
|
|
|
|
TABLE |
| CollExchangeType_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationCategory_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationDateCategory_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationQualifier_Enum |
User |
|
|
|
|
TABLE |
| CollLabelTranscriptionState_Enum |
User |
|
|
|
|
TABLE |
| CollLabelType_Enum |
User |
|
|
|
|
TABLE |
| CollMaterialCategory_Enum |
User |
|
|
|
|
TABLE |
| CollRetrievalType_Enum |
User |
|
|
|
|
TABLE |
| CollSpecimenImageType_Enum |
User |
|
|
|
|
TABLE |
| CollSpecimenRelationType_Enum |
User |
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
User |
|
|
|
|
TABLE |
| CollTaxonomicGroup_Enum |
User |
|
|
|
|
TABLE |
| CollTransactionType_Enum |
User |
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
User |
|
|
|
|
TABLE |
| CollUnitRelationType_Enum |
User |
|
|
|
|
TABLE |
| Entity |
User |
|
|
|
|
TABLE |
| EntityAccessibility_Enum |
User |
|
|
|
|
TABLE |
| EntityContext_Enum |
User |
|
|
|
|
TABLE |
| EntityDetermination_Enum |
User |
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
User |
|
|
|
|
TABLE |
| EntityRepresentation |
User |
|
|
|
|
TABLE |
| EntityUsage |
User |
|
|
|
|
TABLE |
| EntityUsage_Enum |
User |
|
|
|
|
TABLE |
| EntityVisibility_Enum |
User |
|
|
|
|
TABLE |
| ExternalIdentifier |
User |
|
|
|
|
TABLE |
| ExternalIdentifier_log |
|
|
|
|
|
TABLE |
| ExternalIdentifierType |
User |
|
|
|
|
TABLE |
| ExternalIdentifierType_log |
|
|
|
|
|
TABLE |
| Identification |
User |
|
|
|
|
TABLE |
| Identification_log |
|
|
|
|
|
TABLE |
| IdentificationUnit |
User |
|
|
|
|
TABLE |
| IdentificationUnit_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis |
User |
• |
• |
• |
|
TABLE |
| IdentificationUnitAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
User |
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
User |
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis |
User |
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart |
User |
|
• |
• |
|
TABLE |
| IdentificationUnitInPart_log |
|
|
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
User |
|
|
|
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
User |
|
|
|
|
TABLE |
| Method_log |
|
|
|
|
|
TABLE |
| MethodForAnalysis |
User |
|
|
|
|
TABLE |
| MethodForProcessing |
User |
|
|
|
|
TABLE |
| Parameter |
User |
|
|
|
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
User |
|
|
|
|
TABLE |
| Processing |
User |
|
|
|
|
TABLE |
| Processing_log |
|
|
|
|
|
TABLE |
| ProcessingMaterialCategory |
User |
|
|
|
|
TABLE |
| ProjectAnalysis |
User |
|
|
|
|
TABLE |
| ProjectMaterialCategory |
User |
|
|
|
|
TABLE |
| ProjectProcessing |
User |
|
|
|
|
TABLE |
| ProjectTaxonomicGroup |
User |
|
|
|
|
TABLE |
| ProjectUser |
User |
|
|
|
|
TABLE |
| Property |
User |
|
|
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
User |
|
|
|
|
TABLE |
| ReplicationPublisher |
|
|
|
|
|
TABLE |
| Task |
User |
|
|
|
|
TABLE |
| Task_log |
|
|
|
|
|
TABLE |
| TaskDateType_Enum |
User |
|
|
|
|
TABLE |
| TaskModule |
User |
|
|
|
|
TABLE |
| TaskModule_log |
|
|
|
|
|
TABLE |
| TaskModuleType_Enum |
User |
|
|
|
|
TABLE |
| TaskResult |
User |
|
|
|
|
TABLE |
| TaskResult_log |
|
|
|
|
|
TABLE |
| TaskType_Enum |
User |
|
|
|
|
TABLE |
| Transaction |
User |
|
|
|
|
TABLE |
| Transaction_log |
|
|
|
|
|
TABLE |
| TransactionAgent |
|
|
|
|
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
User |
|
|
|
|
TABLE |
| TransactionDocument |
User |
|
|
|
|
TABLE |
| TransactionDocument_log |
|
|
|
|
|
TABLE |
| TransactionPayment |
|
|
|
|
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
User |
|
|
|
|
VIEW |
| AnnotationPart |
User |
|
|
|
|
VIEW |
| AnnotationSpecimen |
User |
|
|
|
|
VIEW |
| AnnotationUnit |
User |
|
|
|
|
VIEW |
| CollectionAgent_Core |
User |
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
User |
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
User |
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
User |
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
|
|
|
|
|
VIEW |
| FirstLinesIdentification |
User |
|
|
|
|
VIEW |
| Identification_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
User |
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
User |
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
User |
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
User |
|
|
|
|
VIEW |
| TransactionList_H7 |
User |
|
|
|
|
VIEW |
| TransactionPermit |
User |
|
|
|
|
VIEW |
| TransactionRegulation |
User |
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
User |
|
|
|
|
VIEW |
| ViewCollectionEventImage |
User |
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
User |
|
|
|
|
VIEW |
| ViewCollectionImage |
User |
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
User |
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
User |
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
User |
|
|
|
|
VIEW |
| AgentOneString |
|
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
User |
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
User |
|
|
|
|
FUNCTION |
| AnalysisList |
User |
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
User |
|
|
|
|
FUNCTION |
| AnalysisProjectList |
User |
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
User |
FUNCTION |
| CollCharacterType_List |
User |
|
|
|
|
FUNCTION |
| CollDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollectionChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
User |
|
|
|
|
FUNCTION |
| CollectionLocation |
User |
|
|
|
|
FUNCTION |
| CollectionLocationAll |
User |
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
User |
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
User |
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
User |
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollEventImageType_List |
User |
|
|
|
|
FUNCTION |
| CollExchangeType_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
User |
|
|
|
|
FUNCTION |
| CollLabelType_List |
User |
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
User |
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
User |
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
User |
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
User |
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
User |
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
|
|
|
|
|
FUNCTION |
| CurrentUser |
User |
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
User |
FUNCTION |
| DefaultProjectID |
|
|
|
|
User |
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
User |
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
User |
FUNCTION |
| EntityInformation_2 |
User |
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
User |
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
User |
FUNCTION |
| EventSeriesChildNodes |
User |
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
User |
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
User |
FUNCTION |
| EventSuperiorList |
User |
|
|
|
|
FUNCTION |
| FirstLines_4 |
User |
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesSeries |
User |
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
User |
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
User |
|
|
|
|
FUNCTION |
| ManagerCollectionList |
|
|
|
|
|
FUNCTION |
| MethodChildNodes |
User |
|
|
|
|
FUNCTION |
| MethodHierarchy |
User |
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
User |
|
|
|
|
FUNCTION |
| NameList |
User |
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
User |
|
|
|
|
FUNCTION |
| NameListPlants |
User |
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
|
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
|
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
User |
FUNCTION |
| ProcessingChildNodes |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
User |
|
|
|
|
FUNCTION |
| ProcessingListForPart |
User |
|
|
|
|
FUNCTION |
| ProcessingProjectList |
User |
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
User |
FUNCTION |
| RequesterCollectionList |
|
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
User |
FUNCTION |
| TaskChildNodes |
User |
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaskHierarchy |
User |
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
User |
FUNCTION |
| TransactionChildNodes |
User |
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
User |
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
User |
FUNCTION |
| TransactionHierarchy |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| UserCollectionList |
User |
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
User |
FUNCTION |
| Version |
|
|
|
|
User |
FUNCTION |
| VersionClient |
|
|
|
|
User |
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
User |
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
|
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
User |
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
|
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| SetXmlValue |
|
|
|
|
User |
PROCEDURE |
| Inheriting from roles: |
|
|
|
|
|
|
Role TransactionUser
Database Role with reading accesss to Transaction information.
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
|
|
|
|
|
TABLE |
| Analysis_log |
|
|
|
|
|
TABLE |
| AnalysisResult |
|
|
|
|
|
TABLE |
| AnalysisTaxonomicGroup |
|
|
|
|
|
TABLE |
| Annotation |
|
|
|
|
|
TABLE |
| Annotation_log |
|
|
|
|
|
TABLE |
| AnnotationType_Enum |
|
|
|
|
|
TABLE |
| AnonymCollector |
|
|
|
|
|
TABLE |
| CacheDatabase2 |
|
|
|
|
|
TABLE |
| CacheDescription |
|
|
|
|
|
TABLE |
| CollCircumstances_Enum |
|
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
|
|
|
|
|
TABLE |
| CollCollectionType_Enum |
|
|
|
|
|
TABLE |
| CollDateCategory_Enum |
|
|
|
|
|
TABLE |
| Collection |
|
|
|
|
|
TABLE |
| Collection_log |
|
|
|
|
|
TABLE |
| CollectionAgent |
|
|
|
|
|
TABLE |
| CollectionAgent_log |
|
|
|
|
|
TABLE |
| CollectionEvent |
|
|
|
|
|
TABLE |
| CollectionEvent_log |
|
|
|
|
|
TABLE |
| CollectionEventImage |
|
|
|
|
|
TABLE |
| CollectionEventImage_log |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation_log |
|
|
|
|
|
TABLE |
| CollectionEventMethod |
|
|
|
|
|
TABLE |
| CollectionEventMethod_log |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue_log |
|
|
|
|
|
TABLE |
| CollectionEventProperty |
|
|
|
|
|
TABLE |
| CollectionEventProperty_log |
|
|
|
|
|
TABLE |
| CollectionEventRegulation |
|
|
|
|
|
TABLE |
| CollectionEventRegulation_log |
|
|
|
|
|
TABLE |
| CollectionEventSeries |
|
|
|
|
|
TABLE |
| CollectionEventSeries_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
|
|
|
|
|
TABLE |
| CollectionImage_log |
|
|
|
|
|
TABLE |
| CollectionManager |
|
|
|
|
|
TABLE |
| CollectionProject |
|
|
|
|
|
TABLE |
| CollectionProject_log |
|
|
|
|
|
TABLE |
| CollectionSpecimen |
|
|
|
|
|
TABLE |
| CollectionSpecimen_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction |
• |
|
|
|
|
TABLE |
| CollectionSpecimenTransaction_log |
|
|
|
|
|
TABLE |
| CollectionTask |
|
|
|
|
|
TABLE |
| CollectionTask_log |
|
|
|
|
|
TABLE |
| CollectionTaskImage |
|
|
|
|
|
TABLE |
| CollectionTaskImage_log |
|
|
|
|
|
TABLE |
| CollectionTaskMetric |
|
|
|
|
|
TABLE |
| CollectionTaskMetric_log |
|
|
|
|
|
TABLE |
| CollEventDateCategory_Enum |
|
|
|
|
|
TABLE |
| CollEventImageType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
|
|
|
|
|
TABLE |
| CollExchangeType_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationCategory_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationDateCategory_Enum |
|
|
|
|
|
TABLE |
| CollIdentificationQualifier_Enum |
|
|
|
|
|
TABLE |
| CollLabelTranscriptionState_Enum |
|
|
|
|
|
TABLE |
| CollLabelType_Enum |
|
|
|
|
|
TABLE |
| CollMaterialCategory_Enum |
|
|
|
|
|
TABLE |
| CollRetrievalType_Enum |
|
|
|
|
|
TABLE |
| CollSpecimenImageType_Enum |
|
|
|
|
|
TABLE |
| CollSpecimenRelationType_Enum |
|
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
|
|
|
|
|
TABLE |
| CollTaxonomicGroup_Enum |
|
|
|
|
|
TABLE |
| CollTransactionType_Enum |
|
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
|
|
|
|
|
TABLE |
| CollUnitRelationType_Enum |
|
|
|
|
|
TABLE |
| Entity |
|
|
|
|
|
TABLE |
| EntityAccessibility_Enum |
|
|
|
|
|
TABLE |
| EntityContext_Enum |
|
|
|
|
|
TABLE |
| EntityDetermination_Enum |
|
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
|
|
|
|
|
TABLE |
| EntityRepresentation |
|
|
|
|
|
TABLE |
| EntityUsage |
|
|
|
|
|
TABLE |
| EntityUsage_Enum |
|
|
|
|
|
TABLE |
| EntityVisibility_Enum |
|
|
|
|
|
TABLE |
| ExternalIdentifier |
|
|
|
|
|
TABLE |
| ExternalIdentifier_log |
|
|
|
|
|
TABLE |
| ExternalIdentifierType |
|
|
|
|
|
TABLE |
| ExternalIdentifierType_log |
|
|
|
|
|
TABLE |
| Identification |
|
|
|
|
|
TABLE |
| Identification_log |
|
|
|
|
|
TABLE |
| IdentificationUnit |
|
|
|
|
|
TABLE |
| IdentificationUnit_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart_log |
|
|
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
|
|
|
|
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
|
|
|
|
|
TABLE |
| Method_log |
|
|
|
|
|
TABLE |
| MethodForAnalysis |
|
|
|
|
|
TABLE |
| MethodForProcessing |
|
|
|
|
|
TABLE |
| Parameter |
|
|
|
|
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
|
|
|
|
|
TABLE |
| Processing |
|
|
|
|
|
TABLE |
| Processing_log |
|
|
|
|
|
TABLE |
| ProcessingMaterialCategory |
|
|
|
|
|
TABLE |
| ProjectAnalysis |
|
|
|
|
|
TABLE |
| ProjectMaterialCategory |
|
|
|
|
|
TABLE |
| ProjectProcessing |
|
|
|
|
|
TABLE |
| ProjectTaxonomicGroup |
|
|
|
|
|
TABLE |
| ProjectUser |
|
|
|
|
|
TABLE |
| Property |
|
|
|
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
|
|
|
|
|
TABLE |
| ReplicationPublisher |
|
|
|
|
|
TABLE |
| Task |
|
|
|
|
|
TABLE |
| Task_log |
|
|
|
|
|
TABLE |
| TaskDateType_Enum |
|
|
|
|
|
TABLE |
| TaskModule |
|
|
|
|
|
TABLE |
| TaskModule_log |
|
|
|
|
|
TABLE |
| TaskModuleType_Enum |
|
|
|
|
|
TABLE |
| TaskResult |
|
|
|
|
|
TABLE |
| TaskResult_log |
|
|
|
|
|
TABLE |
| TaskType_Enum |
|
|
|
|
|
TABLE |
| Transaction |
• |
|
|
|
|
TABLE |
| Transaction_log |
|
|
|
|
|
TABLE |
| TransactionAgent |
• |
|
|
|
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
|
|
|
|
|
TABLE |
| TransactionDocument |
|
|
|
|
|
TABLE |
| TransactionDocument_log |
|
|
|
|
|
TABLE |
| TransactionPayment |
• |
|
|
|
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
|
|
|
|
|
VIEW |
| AnnotationPart |
|
|
|
|
|
VIEW |
| AnnotationSpecimen |
|
|
|
|
|
VIEW |
| AnnotationUnit |
|
|
|
|
|
VIEW |
| CollectionAgent_Core |
|
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
|
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
|
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
|
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
|
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
|
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
|
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
|
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
|
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
|
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
|
|
|
|
|
VIEW |
| FirstLinesIdentification |
|
|
|
|
|
VIEW |
| Identification_Core2 |
|
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
|
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
|
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
|
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
|
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
|
|
|
|
|
VIEW |
| TransactionList_H7 |
|
|
|
|
|
VIEW |
| TransactionPermit |
|
|
|
|
|
VIEW |
| TransactionRegulation |
|
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
|
|
|
|
|
VIEW |
| ViewCollectionEventImage |
|
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
|
|
|
|
|
VIEW |
| ViewCollectionImage |
|
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
|
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
|
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
|
|
|
|
|
VIEW |
| AgentOneString |
|
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
|
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
|
|
|
|
|
FUNCTION |
| AnalysisList |
|
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
|
|
|
|
|
FUNCTION |
| AnalysisProjectList |
|
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
|
FUNCTION |
| CollCharacterType_List |
|
|
|
|
|
FUNCTION |
| CollDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollectionChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
|
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
|
|
|
|
|
FUNCTION |
| CollectionLocation |
|
|
|
|
|
FUNCTION |
| CollectionLocationAll |
|
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
|
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
|
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
|
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
|
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
|
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
|
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
|
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollEventImageType_List |
|
|
|
|
|
FUNCTION |
| CollExchangeType_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
|
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
|
|
|
|
|
FUNCTION |
| CollLabelType_List |
|
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
|
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
|
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
|
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
|
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
|
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
|
|
|
|
|
FUNCTION |
| CurrentUser |
|
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
|
FUNCTION |
| DefaultProjectID |
|
|
|
|
|
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
|
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
|
FUNCTION |
| EntityInformation_2 |
|
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
|
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
|
FUNCTION |
| EventSeriesChildNodes |
|
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
|
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
|
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
|
FUNCTION |
| EventSuperiorList |
|
|
|
|
|
FUNCTION |
| FirstLines_4 |
|
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
|
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
|
|
|
|
|
FUNCTION |
| FirstLinesSeries |
|
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
|
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
|
|
|
|
|
FUNCTION |
| ManagerCollectionList |
|
|
|
|
|
FUNCTION |
| MethodChildNodes |
|
|
|
|
|
FUNCTION |
| MethodHierarchy |
|
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
|
|
|
|
|
FUNCTION |
| NameList |
|
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
|
|
|
|
|
FUNCTION |
| NameListPlants |
|
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
|
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
|
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
|
FUNCTION |
| ProcessingChildNodes |
|
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
|
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
|
|
|
|
|
FUNCTION |
| ProcessingListForPart |
|
|
|
|
|
FUNCTION |
| ProcessingProjectList |
|
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
|
FUNCTION |
| RequesterCollectionList |
|
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
|
FUNCTION |
| TaskChildNodes |
|
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
|
FUNCTION |
| TaskHierarchy |
|
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
|
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
|
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
|
FUNCTION |
| TransactionChildNodes |
|
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
|
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
|
FUNCTION |
| TransactionHierarchy |
|
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
|
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
|
|
|
|
|
FUNCTION |
| UserCollectionList |
|
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
|
FUNCTION |
| Version |
|
|
|
|
|
FUNCTION |
| VersionClient |
|
|
|
|
|
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
|
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
|
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
|
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
|
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
|
PROCEDURE |
| SetXmlValue |
|
|
|
|
|
PROCEDURE |
Role Typist
Write access to most objects. Can not delete data and not insert data into tables CollectionReference and CollectionReference
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
User |
|
|
|
|
TABLE |
| Analysis_log |
|
|
|
|
|
TABLE |
| AnalysisResult |
User |
|
|
|
|
TABLE |
| AnalysisTaxonomicGroup |
User |
|
|
|
|
TABLE |
| Annotation |
User |
User |
User |
|
|
TABLE |
| Annotation_log |
|
|
|
|
|
TABLE |
| AnnotationType_Enum |
User |
|
|
|
|
TABLE |
| AnonymCollector |
|
|
|
|
|
TABLE |
| CacheDatabase2 |
|
|
|
|
|
TABLE |
| CacheDescription |
User |
User |
User |
User |
|
TABLE |
| CollCircumstances_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
User |
|
|
|
|
TABLE |
| CollCollectionType_Enum |
User |
|
|
|
|
TABLE |
| CollDateCategory_Enum |
User |
|
|
|
|
TABLE |
| Collection |
User |
|
|
|
|
TABLE |
| Collection_log |
|
|
|
|
|
TABLE |
| CollectionAgent |
User |
• |
• |
|
|
TABLE |
| CollectionAgent_log |
• |
• |
|
|
|
TABLE |
| CollectionEvent |
User |
|
• |
|
|
TABLE |
| CollectionEvent_log |
• |
• |
|
|
|
TABLE |
| CollectionEventImage |
User |
• |
• |
|
|
TABLE |
| CollectionEventImage_log |
• |
• |
|
|
|
TABLE |
| CollectionEventLocalisation |
User |
• |
• |
|
|
TABLE |
| CollectionEventLocalisation_log |
• |
• |
|
|
|
TABLE |
| CollectionEventMethod |
User |
|
|
|
|
TABLE |
| CollectionEventMethod_log |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue |
User |
|
|
|
|
TABLE |
| CollectionEventParameterValue_log |
|
|
|
|
|
TABLE |
| CollectionEventProperty |
User |
• |
• |
|
|
TABLE |
| CollectionEventProperty_log |
• |
• |
|
|
|
TABLE |
| CollectionEventRegulation |
User |
|
|
|
|
TABLE |
| CollectionEventRegulation_log |
|
|
|
|
|
TABLE |
| CollectionEventSeries |
User |
|
• |
|
|
TABLE |
| CollectionEventSeries_log |
• |
• |
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
User |
|
• |
|
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
User |
|
|
|
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
User |
|
|
|
|
TABLE |
| CollectionImage_log |
|
|
|
|
|
TABLE |
| CollectionManager |
|
|
|
|
|
TABLE |
| CollectionProject |
User |
|
|
|
|
TABLE |
| CollectionProject_log |
|
|
|
|
|
TABLE |
| CollectionSpecimen |
• |
|
• |
|
|
TABLE |
| CollectionSpecimen_log |
• |
• |
|
|
|
TABLE |
| CollectionSpecimenImage |
• |
• |
|
|
|
TABLE |
| CollectionSpecimenImage_log |
• |
• |
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
User |
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart |
User |
• |
• |
|
|
TABLE |
| CollectionSpecimenPart_log |
• |
• |
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
User |
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing |
User |
• |
• |
|
|
TABLE |
| CollectionSpecimenProcessing_log |
• |
• |
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
User |
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
User |
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
User |
|
|
|
|
TABLE |
| CollectionSpecimenReference_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation |
User |
|
|
|
|
TABLE |
| CollectionSpecimenRelation_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction |
User |
• |
• |
|
|
TABLE |
| CollectionSpecimenTransaction_log |
• |
• |
|
|
|
TABLE |
| CollectionTask |
User |
|
|
|
|
TABLE |
| CollectionTask_log |
|
|
|
|
|
TABLE |
| CollectionTaskImage |
User |
|
|
|
|
TABLE |
| CollectionTaskImage_log |
|
|
|
|
|
TABLE |
| CollectionTaskMetric |
User |
|
|
|
|
TABLE |
| CollectionTaskMetric_log |
|
|
|
|
|
TABLE |
| CollEventDateCategory_Enum |
User |
|
|
|
|
TABLE |
| CollEventImageType_Enum |
User |
|
|
|
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
User |
|
|
|
|
TABLE |
| CollExchangeType_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationCategory_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationDateCategory_Enum |
User |
|
|
|
|
TABLE |
| CollIdentificationQualifier_Enum |
User |
|
|
|
|
TABLE |
| CollLabelTranscriptionState_Enum |
User |
|
|
|
|
TABLE |
| CollLabelType_Enum |
User |
|
|
|
|
TABLE |
| CollMaterialCategory_Enum |
User |
|
|
|
|
TABLE |
| CollRetrievalType_Enum |
User |
|
|
|
|
TABLE |
| CollSpecimenImageType_Enum |
User |
|
|
|
|
TABLE |
| CollSpecimenRelationType_Enum |
User |
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
User |
|
|
|
|
TABLE |
| CollTaxonomicGroup_Enum |
User |
|
|
|
|
TABLE |
| CollTransactionType_Enum |
User |
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
User |
|
|
|
|
TABLE |
| CollUnitRelationType_Enum |
User |
|
|
|
|
TABLE |
| Entity |
User |
|
|
|
|
TABLE |
| EntityAccessibility_Enum |
User |
|
|
|
|
TABLE |
| EntityContext_Enum |
User |
|
|
|
|
TABLE |
| EntityDetermination_Enum |
User |
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
User |
|
|
|
|
TABLE |
| EntityRepresentation |
User |
|
|
|
|
TABLE |
| EntityUsage |
User |
|
|
|
|
TABLE |
| EntityUsage_Enum |
User |
|
|
|
|
TABLE |
| EntityVisibility_Enum |
User |
|
|
|
|
TABLE |
| ExternalIdentifier |
User |
|
|
|
|
TABLE |
| ExternalIdentifier_log |
|
|
|
|
|
TABLE |
| ExternalIdentifierType |
User |
|
|
|
|
TABLE |
| ExternalIdentifierType_log |
|
|
|
|
|
TABLE |
| Identification |
User |
• |
• |
|
|
TABLE |
| Identification_log |
• |
• |
|
|
|
TABLE |
| IdentificationUnit |
User |
• |
• |
|
|
TABLE |
| IdentificationUnit_log |
• |
• |
|
|
|
TABLE |
| IdentificationUnitAnalysis |
User |
• |
• |
|
|
TABLE |
| IdentificationUnitAnalysis_log |
• |
• |
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
User |
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
User |
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis |
User |
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart |
User |
• |
• |
|
|
TABLE |
| IdentificationUnitInPart_log |
• |
• |
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
User |
|
|
|
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
User |
|
|
|
|
TABLE |
| Method_log |
|
|
|
|
|
TABLE |
| MethodForAnalysis |
User |
|
|
|
|
TABLE |
| MethodForProcessing |
User |
|
|
|
|
TABLE |
| Parameter |
User |
|
|
|
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
User |
|
|
|
|
TABLE |
| Processing |
User |
|
|
|
|
TABLE |
| Processing_log |
|
|
|
|
|
TABLE |
| ProcessingMaterialCategory |
User |
|
|
|
|
TABLE |
| ProjectAnalysis |
User |
|
|
|
|
TABLE |
| ProjectMaterialCategory |
User |
|
|
|
|
TABLE |
| ProjectProcessing |
User |
|
|
|
|
TABLE |
| ProjectTaxonomicGroup |
User |
|
|
|
|
TABLE |
| ProjectUser |
User |
|
|
|
|
TABLE |
| Property |
User |
|
|
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
User |
|
|
|
|
TABLE |
| ReplicationPublisher |
|
|
|
|
|
TABLE |
| Task |
User |
|
|
|
|
TABLE |
| Task_log |
|
|
|
|
|
TABLE |
| TaskDateType_Enum |
User |
|
|
|
|
TABLE |
| TaskModule |
User |
|
|
|
|
TABLE |
| TaskModule_log |
|
|
|
|
|
TABLE |
| TaskModuleType_Enum |
User |
|
|
|
|
TABLE |
| TaskResult |
User |
|
|
|
|
TABLE |
| TaskResult_log |
|
|
|
|
|
TABLE |
| TaskType_Enum |
User |
|
|
|
|
TABLE |
| Transaction |
User |
|
|
|
|
TABLE |
| Transaction_log |
• |
• |
|
|
|
TABLE |
| TransactionAgent |
|
|
|
|
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
User |
|
|
|
|
TABLE |
| TransactionDocument |
User |
• |
• |
|
|
TABLE |
| TransactionDocument_log |
• |
• |
|
|
|
TABLE |
| TransactionPayment |
|
|
|
|
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
User |
|
|
|
|
VIEW |
| AnnotationPart |
User |
|
|
|
|
VIEW |
| AnnotationSpecimen |
User |
|
|
|
|
VIEW |
| AnnotationUnit |
User |
|
|
|
|
VIEW |
| CollectionAgent_Core |
User |
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
User |
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
User |
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
User |
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
User |
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
User |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
User |
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
|
|
|
|
|
VIEW |
| FirstLinesIdentification |
User |
|
|
|
|
VIEW |
| Identification_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
User |
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
User |
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
User |
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
User |
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
User |
|
|
|
|
VIEW |
| TransactionList_H7 |
User |
|
|
|
|
VIEW |
| TransactionPermit |
User |
|
|
|
|
VIEW |
| TransactionRegulation |
User |
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
User |
|
|
|
|
VIEW |
| ViewCollectionEventImage |
User |
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
User |
|
|
|
|
VIEW |
| ViewCollectionImage |
User |
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
User |
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
User |
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
User |
|
|
|
|
VIEW |
| AgentOneString |
|
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
User |
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
User |
|
|
|
|
FUNCTION |
| AnalysisList |
User |
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
User |
|
|
|
|
FUNCTION |
| AnalysisProjectList |
User |
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
User |
FUNCTION |
| CollCharacterType_List |
User |
|
|
|
|
FUNCTION |
| CollDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollectionChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
User |
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
User |
|
|
|
|
FUNCTION |
| CollectionLocation |
User |
|
|
|
|
FUNCTION |
| CollectionLocationAll |
User |
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
User |
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
User |
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
User |
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
User |
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
User |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
User |
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollEventImageType_List |
User |
|
|
|
|
FUNCTION |
| CollExchangeType_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
User |
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
User |
|
|
|
|
FUNCTION |
| CollLabelType_List |
User |
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
User |
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
User |
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
User |
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
User |
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
User |
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
|
|
|
|
|
FUNCTION |
| CurrentUser |
User |
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
User |
FUNCTION |
| DefaultProjectID |
|
|
|
|
User |
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
User |
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
User |
FUNCTION |
| EntityInformation_2 |
User |
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
User |
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
User |
FUNCTION |
| EventSeriesChildNodes |
User |
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
User |
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
User |
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
User |
FUNCTION |
| EventSuperiorList |
User |
|
|
|
|
FUNCTION |
| FirstLines_4 |
User |
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
User |
|
|
|
|
FUNCTION |
| FirstLinesSeries |
User |
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
User |
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
User |
|
|
|
|
FUNCTION |
| ManagerCollectionList |
|
|
|
|
|
FUNCTION |
| MethodChildNodes |
User |
|
|
|
|
FUNCTION |
| MethodHierarchy |
User |
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
User |
|
|
|
|
FUNCTION |
| NameList |
User |
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
User |
|
|
|
|
FUNCTION |
| NameListPlants |
User |
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
• |
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
|
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
User |
FUNCTION |
| ProcessingChildNodes |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
User |
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
User |
|
|
|
|
FUNCTION |
| ProcessingListForPart |
User |
|
|
|
|
FUNCTION |
| ProcessingProjectList |
User |
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
User |
FUNCTION |
| RequesterCollectionList |
|
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
User |
FUNCTION |
| TaskChildNodes |
User |
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaskHierarchy |
User |
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
User |
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
User |
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
User |
FUNCTION |
| TransactionChildNodes |
User |
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
User |
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
User |
FUNCTION |
| TransactionHierarchy |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
User |
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
User |
|
|
|
|
FUNCTION |
| UserCollectionList |
User |
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
User |
FUNCTION |
| Version |
|
|
|
|
User |
FUNCTION |
| VersionClient |
|
|
|
|
User |
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
User |
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
|
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
User |
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
|
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
User |
PROCEDURE |
| SetXmlValue |
|
|
|
|
User |
PROCEDURE |
| Inheriting from roles: |
|
|
|
|
|
|
Role User
Restricted to read access to all tables. Can insert data into table Annotation
| Permissions |
SELECT |
INSERT |
UPDATE |
DELETE |
EXECUTE |
Type |
| Analysis |
• |
|
|
|
|
TABLE |
| Analysis_log |
|
|
|
|
|
TABLE |
| AnalysisResult |
• |
|
|
|
|
TABLE |
| AnalysisTaxonomicGroup |
• |
|
|
|
|
TABLE |
| Annotation |
• |
• |
• |
|
|
TABLE |
| Annotation_log |
|
|
|
|
|
TABLE |
| AnnotationType_Enum |
• |
|
|
|
|
TABLE |
| AnonymCollector |
|
|
|
|
|
TABLE |
| CacheDatabase2 |
|
|
|
|
|
TABLE |
| CacheDescription |
• |
• |
• |
• |
|
TABLE |
| CollCircumstances_Enum |
• |
|
|
|
|
TABLE |
| CollCollectionImageType_Enum |
• |
|
|
|
|
TABLE |
| CollCollectionType_Enum |
• |
|
|
|
|
TABLE |
| CollDateCategory_Enum |
• |
|
|
|
|
TABLE |
| Collection |
• |
|
|
|
|
TABLE |
| Collection_log |
|
|
|
|
|
TABLE |
| CollectionAgent |
• |
|
|
|
|
TABLE |
| CollectionAgent_log |
|
|
|
|
|
TABLE |
| CollectionEvent |
• |
|
|
|
|
TABLE |
| CollectionEvent_log |
|
|
|
|
|
TABLE |
| CollectionEventImage |
• |
|
|
|
|
TABLE |
| CollectionEventImage_log |
|
|
|
|
|
TABLE |
| CollectionEventLocalisation |
• |
|
|
|
|
TABLE |
| CollectionEventLocalisation_log |
|
|
|
|
|
TABLE |
| CollectionEventMethod |
• |
|
|
|
|
TABLE |
| CollectionEventMethod_log |
|
|
|
|
|
TABLE |
| CollectionEventParameterValue |
• |
|
|
|
|
TABLE |
| CollectionEventParameterValue_log |
|
|
|
|
|
TABLE |
| CollectionEventProperty |
• |
|
|
|
|
TABLE |
| CollectionEventProperty_log |
|
|
|
|
|
TABLE |
| CollectionEventRegulation |
• |
|
|
|
|
TABLE |
| CollectionEventRegulation_log |
|
|
|
|
|
TABLE |
| CollectionEventSeries |
• |
|
|
|
|
TABLE |
| CollectionEventSeries_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor |
|
|
|
|
|
TABLE |
| CollectionEventSeriesDescriptor_log |
|
|
|
|
|
TABLE |
| CollectionEventSeriesImage |
• |
|
|
|
|
TABLE |
| CollectionEventSeriesImage_log |
|
|
|
|
|
TABLE |
| CollectionExternalDatasource |
• |
|
|
|
|
TABLE |
| CollectionExternalDatasource_log |
|
|
|
|
|
TABLE |
| CollectionImage |
• |
|
|
|
|
TABLE |
| CollectionImage_log |
|
|
|
|
|
TABLE |
| CollectionManager |
|
|
|
|
|
TABLE |
| CollectionProject |
• |
|
|
|
|
TABLE |
| CollectionProject_log |
|
|
|
|
|
TABLE |
| CollectionSpecimen |
• |
|
|
|
|
TABLE |
| CollectionSpecimen_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImage |
• |
|
|
|
|
TABLE |
| CollectionSpecimenImage_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty |
• |
|
|
|
|
TABLE |
| CollectionSpecimenImageProperty_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPart |
• |
|
|
|
|
TABLE |
| CollectionSpecimenPart_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription |
• |
|
|
|
|
TABLE |
| CollectionSpecimenPartDescription_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessing |
• |
|
|
|
|
TABLE |
| CollectionSpecimenProcessing_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod |
• |
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethod_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter |
• |
|
|
|
|
TABLE |
| CollectionSpecimenProcessingMethodParameter_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenReference |
• |
|
|
|
|
TABLE |
| CollectionSpecimenReference_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenRelation |
• |
|
|
|
|
TABLE |
| CollectionSpecimenRelation_log |
|
|
|
|
|
TABLE |
| CollectionSpecimenTransaction |
• |
|
|
|
|
TABLE |
| CollectionSpecimenTransaction_log |
|
|
|
|
|
TABLE |
| CollectionTask |
• |
|
|
|
|
TABLE |
| CollectionTask_log |
|
|
|
|
|
TABLE |
| CollectionTaskImage |
• |
|
|
|
|
TABLE |
| CollectionTaskImage_log |
|
|
|
|
|
TABLE |
| CollectionTaskMetric |
• |
|
|
|
|
TABLE |
| CollectionTaskMetric_log |
|
|
|
|
|
TABLE |
| CollEventDateCategory_Enum |
• |
|
|
|
|
TABLE |
| CollEventImageType_Enum |
• |
|
|
|
|
TABLE |
| CollEventSeriesDescriptorType_Enum |
|
|
|
|
|
TABLE |
| CollEventSeriesImageType_Enum |
• |
|
|
|
|
TABLE |
| CollExchangeType_Enum |
• |
|
|
|
|
TABLE |
| CollIdentificationCategory_Enum |
• |
|
|
|
|
TABLE |
| CollIdentificationDateCategory_Enum |
• |
|
|
|
|
TABLE |
| CollIdentificationQualifier_Enum |
• |
|
|
|
|
TABLE |
| CollLabelTranscriptionState_Enum |
• |
|
|
|
|
TABLE |
| CollLabelType_Enum |
• |
|
|
|
|
TABLE |
| CollMaterialCategory_Enum |
• |
|
|
|
|
TABLE |
| CollRetrievalType_Enum |
• |
|
|
|
|
TABLE |
| CollSpecimenImageType_Enum |
• |
|
|
|
|
TABLE |
| CollSpecimenRelationType_Enum |
• |
|
|
|
|
TABLE |
| CollTaskMetricAggregation_Enum |
• |
|
|
|
|
TABLE |
| CollTaxonomicGroup_Enum |
• |
|
|
|
|
TABLE |
| CollTransactionType_Enum |
• |
|
|
|
|
TABLE |
| CollTypeStatus_Enum |
• |
|
|
|
|
TABLE |
| CollUnitRelationType_Enum |
• |
|
|
|
|
TABLE |
| Entity |
• |
|
|
|
|
TABLE |
| EntityAccessibility_Enum |
• |
|
|
|
|
TABLE |
| EntityContext_Enum |
• |
|
|
|
|
TABLE |
| EntityDetermination_Enum |
• |
|
|
|
|
TABLE |
| EntityLanguageCode_Enum |
• |
|
|
|
|
TABLE |
| EntityRepresentation |
• |
|
|
|
|
TABLE |
| EntityUsage |
• |
|
|
|
|
TABLE |
| EntityUsage_Enum |
• |
|
|
|
|
TABLE |
| EntityVisibility_Enum |
• |
|
|
|
|
TABLE |
| ExternalIdentifier |
• |
|
|
|
|
TABLE |
| ExternalIdentifier_log |
|
|
|
|
|
TABLE |
| ExternalIdentifierType |
• |
|
|
|
|
TABLE |
| ExternalIdentifierType_log |
|
|
|
|
|
TABLE |
| Identification |
• |
|
|
|
|
TABLE |
| Identification_log |
|
|
|
|
|
TABLE |
| IdentificationUnit |
• |
|
|
|
|
TABLE |
| IdentificationUnit_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysis |
• |
|
|
|
|
TABLE |
| IdentificationUnitAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod |
• |
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethod_log |
|
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter |
• |
|
|
|
|
TABLE |
| IdentificationUnitAnalysisMethodParameter_log |
|
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis |
• |
|
|
|
|
TABLE |
| IdentificationUnitGeoAnalysis_log |
|
|
|
|
|
TABLE |
| IdentificationUnitInPart |
• |
|
|
|
|
TABLE |
| IdentificationUnitInPart_log |
|
|
|
|
|
TABLE |
| LanguageCode_Enum |
|
|
|
|
|
TABLE |
| LocalisationSystem |
• |
|
|
|
|
TABLE |
| MeasurementUnit_Enum |
|
|
|
|
|
TABLE |
| Method |
• |
|
|
|
|
TABLE |
| Method_log |
|
|
|
|
|
TABLE |
| MethodForAnalysis |
• |
|
|
|
|
TABLE |
| MethodForProcessing |
• |
|
|
|
|
TABLE |
| Parameter |
• |
|
|
|
|
TABLE |
| Parameter_log |
|
|
|
|
|
TABLE |
| ParameterValue_Enum |
• |
|
|
|
|
TABLE |
| Processing |
• |
|
|
|
|
TABLE |
| Processing_log |
|
|
|
|
|
TABLE |
| ProcessingMaterialCategory |
• |
|
|
|
|
TABLE |
| ProjectAnalysis |
• |
|
|
|
|
TABLE |
| ProjectMaterialCategory |
• |
|
|
|
|
TABLE |
| ProjectProcessing |
• |
|
|
|
|
TABLE |
| ProjectTaxonomicGroup |
• |
|
|
|
|
TABLE |
| ProjectUser |
• |
|
|
|
|
TABLE |
| Property |
• |
|
|
|
|
TABLE |
| PropertyType_Enum |
|
|
|
|
|
TABLE |
| RegulationType_Enum |
• |
|
|
|
|
TABLE |
| ReplicationPublisher |
|
|
|
|
|
TABLE |
| Task |
• |
|
|
|
|
TABLE |
| Task_log |
|
|
|
|
|
TABLE |
| TaskDateType_Enum |
• |
|
|
|
|
TABLE |
| TaskModule |
• |
|
|
|
|
TABLE |
| TaskModule_log |
|
|
|
|
|
TABLE |
| TaskModuleType_Enum |
• |
|
|
|
|
TABLE |
| TaskResult |
• |
|
|
|
|
TABLE |
| TaskResult_log |
|
|
|
|
|
TABLE |
| TaskType_Enum |
• |
|
|
|
|
TABLE |
| Transaction |
• |
|
|
|
|
TABLE |
| Transaction_log |
|
|
|
|
|
TABLE |
| TransactionAgent |
|
|
|
|
|
TABLE |
| TransactionAgent_log |
|
|
|
|
|
TABLE |
| TransactionComment |
• |
|
|
|
|
TABLE |
| TransactionDocument |
• |
|
|
|
|
TABLE |
| TransactionDocument_log |
|
|
|
|
|
TABLE |
| TransactionPayment |
|
|
|
|
|
TABLE |
| TransactionPayment_log |
|
|
|
|
|
TABLE |
| AnnotationEvent |
• |
|
|
|
|
VIEW |
| AnnotationPart |
• |
|
|
|
|
VIEW |
| AnnotationSpecimen |
• |
|
|
|
|
VIEW |
| AnnotationUnit |
• |
|
|
|
|
VIEW |
| CollectionAgent_Core |
• |
|
|
|
|
VIEW |
| CollectionEvent_Core2 |
• |
|
|
|
|
VIEW |
| CollectionEventID_CanEdit |
• |
|
|
|
|
VIEW |
| CollectionEventID_UserAvailable |
• |
|
|
|
|
VIEW |
| CollectionEventLocalisation_Core |
• |
|
|
|
|
VIEW |
| CollectionSpecimen_Core2 |
• |
|
|
|
|
VIEW |
| CollectionSpecimenID_Available |
• |
|
|
|
|
VIEW |
| CollectionSpecimenID_AvailableReadOnly |
• |
|
|
|
|
VIEW |
| CollectionSpecimenID_CanEdit |
• |
|
|
|
|
VIEW |
| CollectionSpecimenID_Locked |
• |
|
|
|
|
VIEW |
| CollectionSpecimenID_ReadOnly |
• |
|
|
|
|
VIEW |
| CollectionSpecimenID_UserAvailable |
• |
|
|
|
|
VIEW |
| CollectionSpecimenPart_Core2 |
• |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInternal |
• |
|
|
|
|
VIEW |
| CollectionSpecimenRelationInvers |
• |
|
|
|
|
VIEW |
| CollectionSpecimenTransactionRequest |
|
|
|
|
|
VIEW |
| FirstLinesIdentification |
• |
|
|
|
|
VIEW |
| Identification_Core2 |
• |
|
|
|
|
VIEW |
| IdentificationUnit_Core2 |
• |
|
|
|
|
VIEW |
| IdentificationUnitDisplayOrder1 |
• |
|
|
|
|
VIEW |
| ManagerSpecimenPartList |
|
|
|
|
|
VIEW |
| ProjectList |
• |
|
|
|
|
VIEW |
| ProjectListNotReadOnly |
• |
|
|
|
|
VIEW |
| RequesterSpecimenPartList |
|
|
|
|
|
VIEW |
| TransactionForeignRequest |
|
|
|
|
|
VIEW |
| TransactionList |
• |
|
|
|
|
VIEW |
| TransactionList_H7 |
• |
|
|
|
|
VIEW |
| TransactionPermit |
• |
|
|
|
|
VIEW |
| TransactionRegulation |
• |
|
|
|
|
VIEW |
| TransactionRequest |
|
|
|
|
|
VIEW |
| TransactionUserRequest |
|
|
|
|
|
VIEW |
| UserGroups |
|
|
|
|
|
VIEW |
| ViewBaseURL |
• |
|
|
|
|
VIEW |
| ViewCollectionEventImage |
• |
|
|
|
|
VIEW |
| ViewCollectionEventSeriesImage |
• |
|
|
|
|
VIEW |
| ViewCollectionImage |
• |
|
|
|
|
VIEW |
| ViewCollectionSpecimenImage |
• |
|
|
|
|
VIEW |
| ViewDiversityWorkbenchModule |
• |
|
|
|
|
VIEW |
| ViewIdentificationUnitGeoAnalysis |
• |
|
|
|
|
VIEW |
| AgentOneString |
|
|
|
|
|
FUNCTION |
| AnalysisChildNodes |
• |
|
|
|
|
FUNCTION |
| AnalysisHierarchyAll |
• |
|
|
|
|
FUNCTION |
| AnalysisList |
• |
|
|
|
|
FUNCTION |
| AnalysisListForUnit |
• |
|
|
|
|
FUNCTION |
| AnalysisProjectList |
• |
|
|
|
|
FUNCTION |
| AnalysisTaxonomicGroupForProject |
|
|
|
|
|
FUNCTION |
| AverageAltitude |
|
|
|
|
|
FUNCTION |
| BaseURL |
|
|
|
|
• |
FUNCTION |
| CollCharacterType_List |
• |
|
|
|
|
FUNCTION |
| CollDateCategory_List |
• |
|
|
|
|
FUNCTION |
| CollectionChildNodes |
• |
|
|
|
|
FUNCTION |
| CollectionEventSeriesHierarchy |
• |
|
|
|
|
FUNCTION |
| CollectionHierarchy |
• |
|
|
|
|
FUNCTION |
| CollectionHierarchyAll |
• |
|
|
|
|
FUNCTION |
| CollectionHierarchyMulti |
• |
|
|
|
|
FUNCTION |
| CollectionHierarchySuperior |
• |
|
|
|
|
FUNCTION |
| CollectionLocation |
• |
|
|
|
|
FUNCTION |
| CollectionLocationAll |
• |
|
|
|
|
FUNCTION |
| CollectionLocationChildNodes |
• |
|
|
|
|
FUNCTION |
| CollectionLocationMulti |
• |
|
|
|
|
FUNCTION |
| CollectionLocationSuperior |
• |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinateList |
• |
|
|
|
|
FUNCTION |
| CollectionSpecimenCoordinates |
• |
|
|
|
|
FUNCTION |
| CollectionSpecimenRelationInversList |
• |
|
|
|
|
FUNCTION |
| CollectionTaskChildNodes |
• |
|
|
|
|
FUNCTION |
| CollectionTaskCollectionHierarchyAll |
• |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchy |
• |
|
|
|
|
FUNCTION |
| CollectionTaskHierarchyAll |
• |
|
|
|
|
FUNCTION |
| CollectionTaskParentNodes |
• |
|
|
|
|
FUNCTION |
| CollEventDateCategory_List |
• |
|
|
|
|
FUNCTION |
| CollEventImageType_List |
• |
|
|
|
|
FUNCTION |
| CollExchangeType_List |
• |
|
|
|
|
FUNCTION |
| CollIdentificationCategory_List |
• |
|
|
|
|
FUNCTION |
| CollIdentificationDateCategory_List |
• |
|
|
|
|
FUNCTION |
| CollIdentificationQualifier_List |
• |
|
|
|
|
FUNCTION |
| CollLabelType_List |
• |
|
|
|
|
FUNCTION |
| CollMaterialCategory_List |
• |
|
|
|
|
FUNCTION |
| CollSpecimenImageType_List |
• |
|
|
|
|
FUNCTION |
| CollTranscriptionState_List |
• |
|
|
|
|
FUNCTION |
| CollTypeStatus_List |
• |
|
|
|
|
FUNCTION |
| ColTaxonomicGroup_List |
• |
|
|
|
|
FUNCTION |
| CuratorCollectionHierarchyList |
|
|
|
|
|
FUNCTION |
| CurrentUser |
• |
|
|
|
|
FUNCTION |
| CurrentUserName |
|
|
|
|
• |
FUNCTION |
| DefaultProjectID |
|
|
|
|
• |
FUNCTION |
| DiversityCollectionCacheDatabaseName |
|
|
|
|
• |
FUNCTION |
| DiversityWorkbenchModule |
|
|
|
|
• |
FUNCTION |
| EntityInformation_2 |
• |
|
|
|
|
FUNCTION |
| EventDescription |
|
|
|
|
• |
FUNCTION |
| EventDescriptionSuperior |
|
|
|
|
• |
FUNCTION |
| EventSeriesChildNodes |
• |
|
|
|
|
FUNCTION |
| EventSeriesHierarchy |
• |
|
|
|
|
FUNCTION |
| EventSeriesSuperiorList |
• |
|
|
|
|
FUNCTION |
| EventSeriesTopID |
|
|
|
|
|
FUNCTION |
| EventSpecimenNumber |
|
|
|
|
• |
FUNCTION |
| EventSuperiorList |
• |
|
|
|
|
FUNCTION |
| FirstLines_4 |
• |
|
|
|
|
FUNCTION |
| FirstLinesEvent_2 |
• |
|
|
|
|
FUNCTION |
| FirstLinesPart_2 |
• |
|
|
|
|
FUNCTION |
| FirstLinesSeries |
• |
|
|
|
|
FUNCTION |
| FirstLinesUnit_4 |
• |
|
|
|
|
FUNCTION |
| LocalisationSystem_List |
• |
|
|
|
|
FUNCTION |
| ManagerCollectionList |
|
|
|
|
|
FUNCTION |
| MethodChildNodes |
• |
|
|
|
|
FUNCTION |
| MethodHierarchy |
• |
|
|
|
|
FUNCTION |
| MethodHierarchyAll |
• |
|
|
|
|
FUNCTION |
| NameList |
• |
|
|
|
|
FUNCTION |
| NameListMyxomycetes |
• |
|
|
|
|
FUNCTION |
| NameListPlants |
• |
|
|
|
|
FUNCTION |
| NextFreeAccNr |
|
|
|
|
|
FUNCTION |
| NextFreeAccNumber |
|
|
|
|
|
FUNCTION |
| PrivacyConsentInfo |
|
|
|
|
• |
FUNCTION |
| ProcessingChildNodes |
• |
|
|
|
|
FUNCTION |
| ProcessingHierarchy |
• |
|
|
|
|
FUNCTION |
| ProcessingHierarchyAll |
• |
|
|
|
|
FUNCTION |
| ProcessingListForPart |
• |
|
|
|
|
FUNCTION |
| ProcessingProjectList |
• |
|
|
|
|
FUNCTION |
| ProjectDataLastChanges |
|
|
|
|
• |
FUNCTION |
| RequesterCollectionList |
|
|
|
|
|
FUNCTION |
| StableIdentifier |
|
|
|
|
• |
FUNCTION |
| TaskChildNodes |
• |
|
|
|
|
FUNCTION |
| TaskCollectionHierarchySeparator |
|
|
|
|
• |
FUNCTION |
| TaskHierarchy |
• |
|
|
|
|
FUNCTION |
| TaskHierarchyAll |
• |
|
|
|
|
FUNCTION |
| TaskHierarchySeparator |
|
|
|
|
• |
FUNCTION |
| TaxonWithQualifier |
|
|
|
|
• |
FUNCTION |
| TransactionChildNodes |
• |
|
|
|
|
FUNCTION |
| TransactionChildNodesAccess |
• |
|
|
|
|
FUNCTION |
| TransactionCurrency |
|
|
|
|
• |
FUNCTION |
| TransactionHierarchy |
• |
|
|
|
|
FUNCTION |
| TransactionHierarchyAccess |
• |
|
|
|
|
FUNCTION |
| TransactionHierarchyAll |
• |
|
|
|
|
FUNCTION |
| UserCollectionList |
• |
|
|
|
|
FUNCTION |
| UserID |
|
|
|
|
• |
FUNCTION |
| Version |
|
|
|
|
• |
FUNCTION |
| VersionClient |
|
|
|
|
• |
FUNCTION |
| DeleteXmlAttribute |
|
|
|
|
• |
PROCEDURE |
| DeleteXmlNode |
|
|
|
|
• |
PROCEDURE |
| procCopyCollectionSpecimen2 |
|
|
|
|
|
PROCEDURE |
| procCopyCollectionSpecimenPart |
|
|
|
|
|
PROCEDURE |
| procFillCacheDescription |
|
|
|
|
• |
PROCEDURE |
| procInsertCollectionEventCopy |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionEvent |
|
|
|
|
|
PROCEDURE |
| procSetVersionCollectionSpecimen |
|
|
|
|
|
PROCEDURE |
| SetXmlAttribute |
|
|
|
|
• |
PROCEDURE |
| SetXmlValue |
|
|
|
|
• |
PROCEDURE |
Details of the Database
Details for certain domains within the database. For clarity some tables are not included in the main ER-diagram, but are included in the corresonding ER-diagram for the domain.
Subsections of Details
Diversity Collection
Event Tables
The image below shows the tables related to collection event.
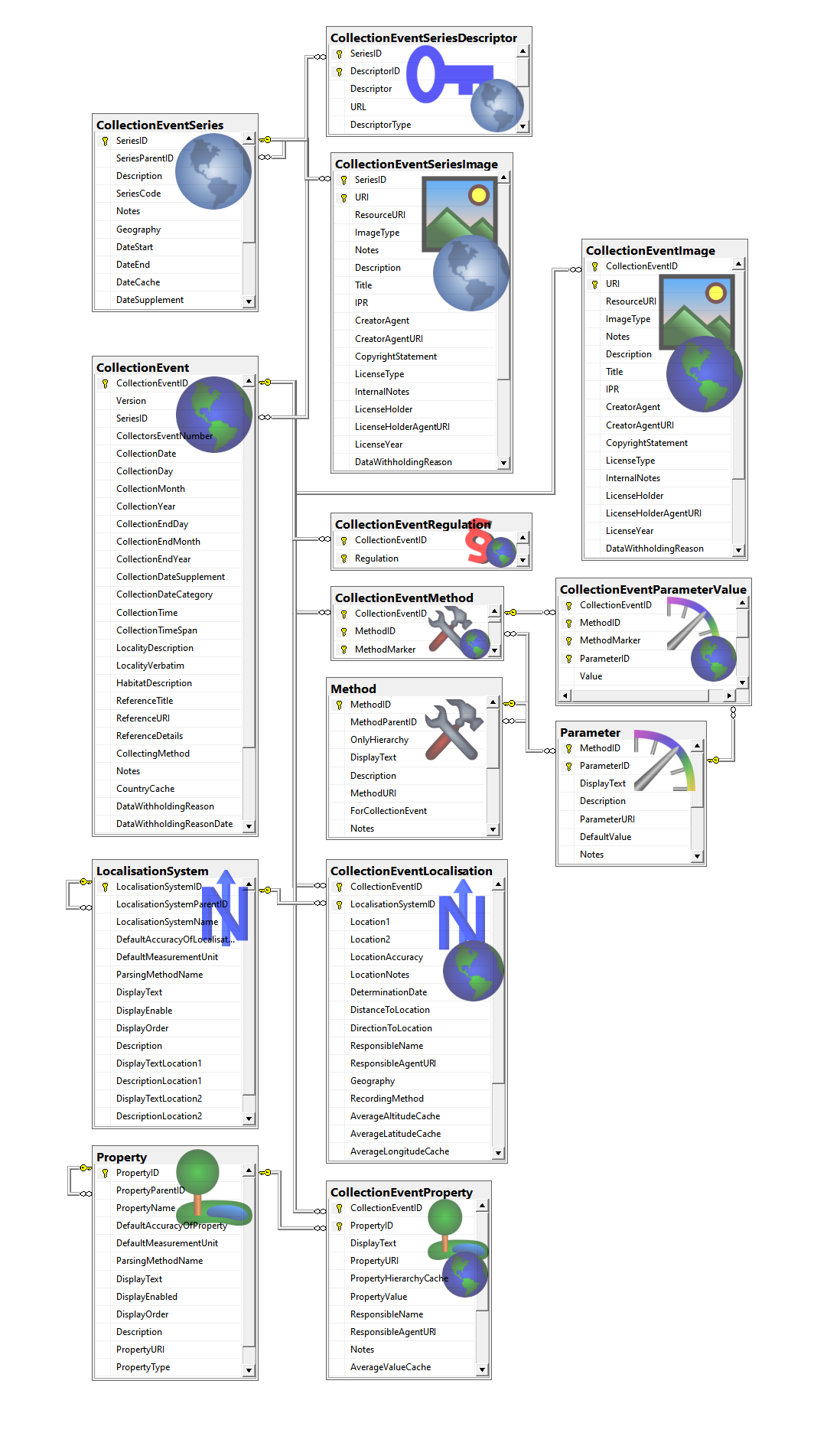
Diversity Collection
Regulation Tables
The image below shows the tables related to the regluation.
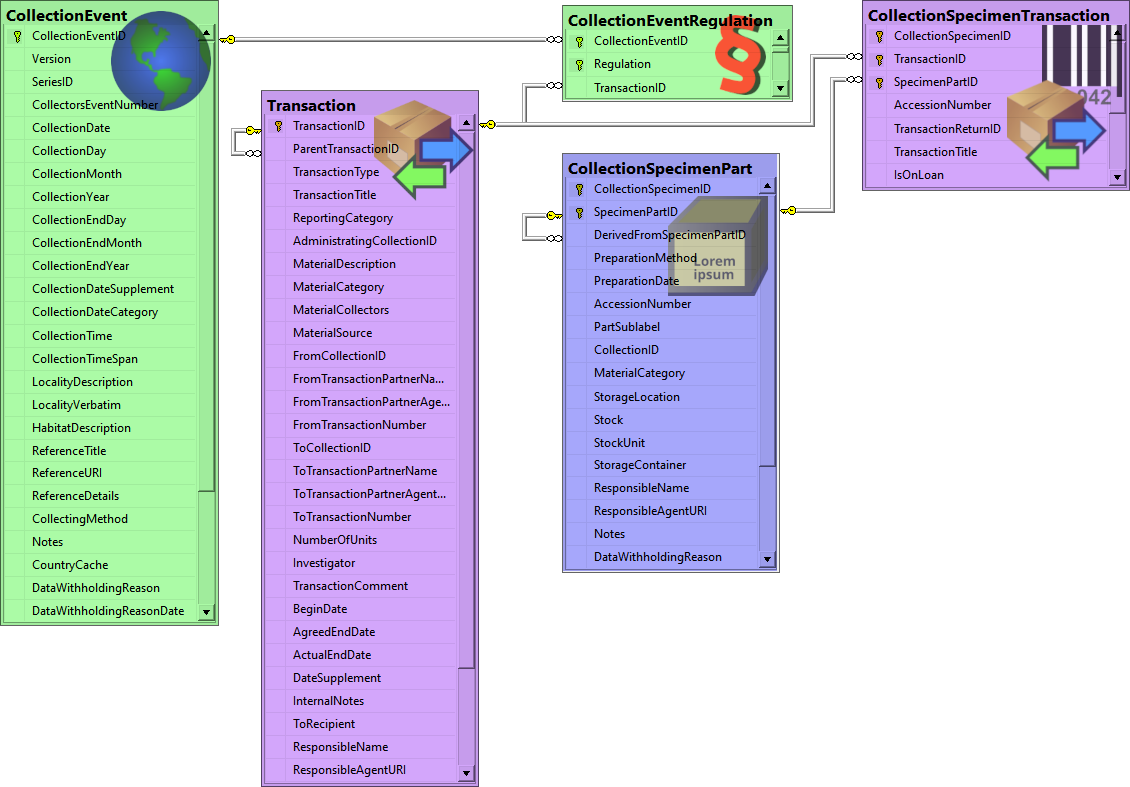
Diversity Collection
Method Tables
The image below shows the tables related to the method.
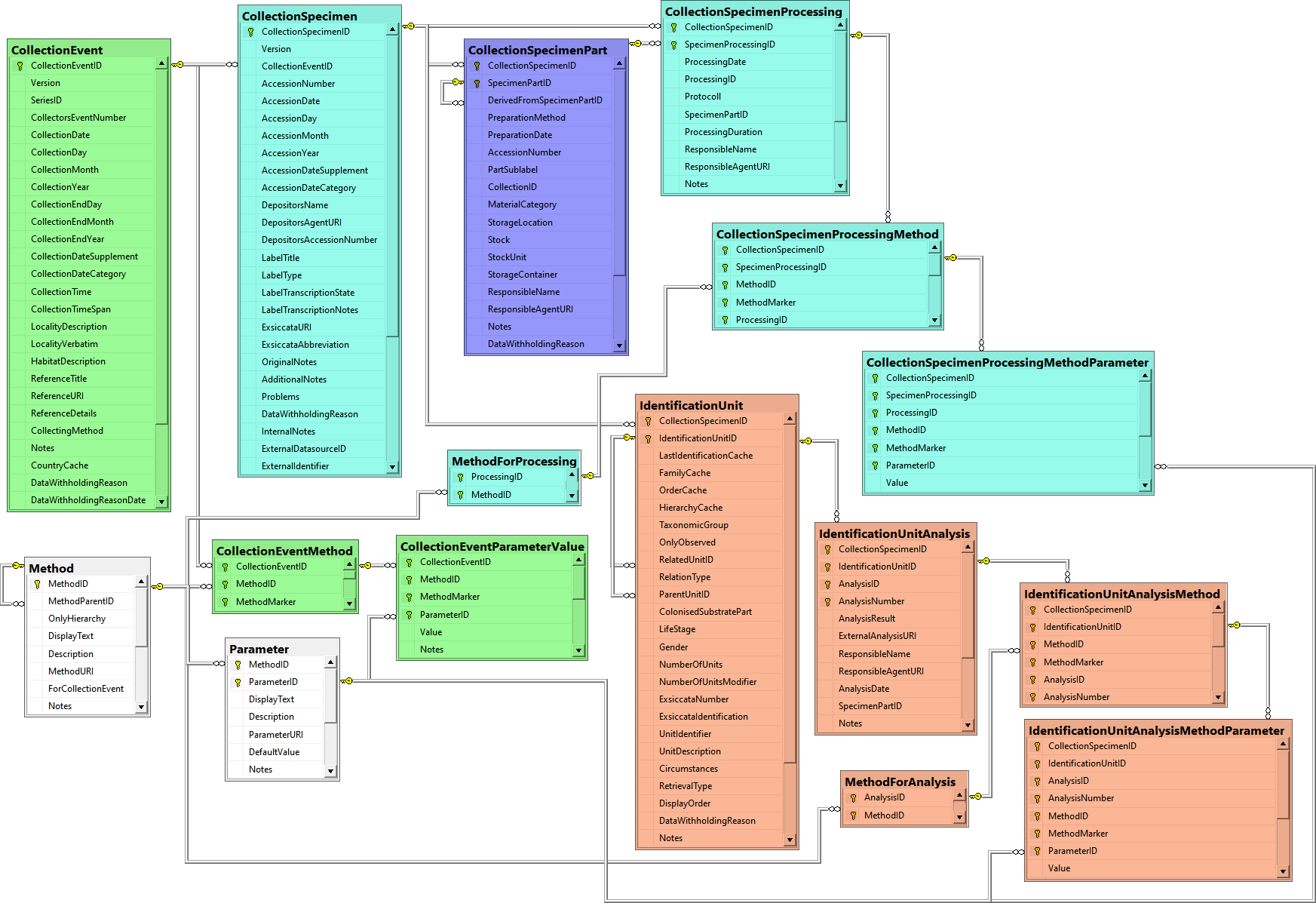
Diversity Collection
Collection Tables
The image below shows the tables related to the Collection.
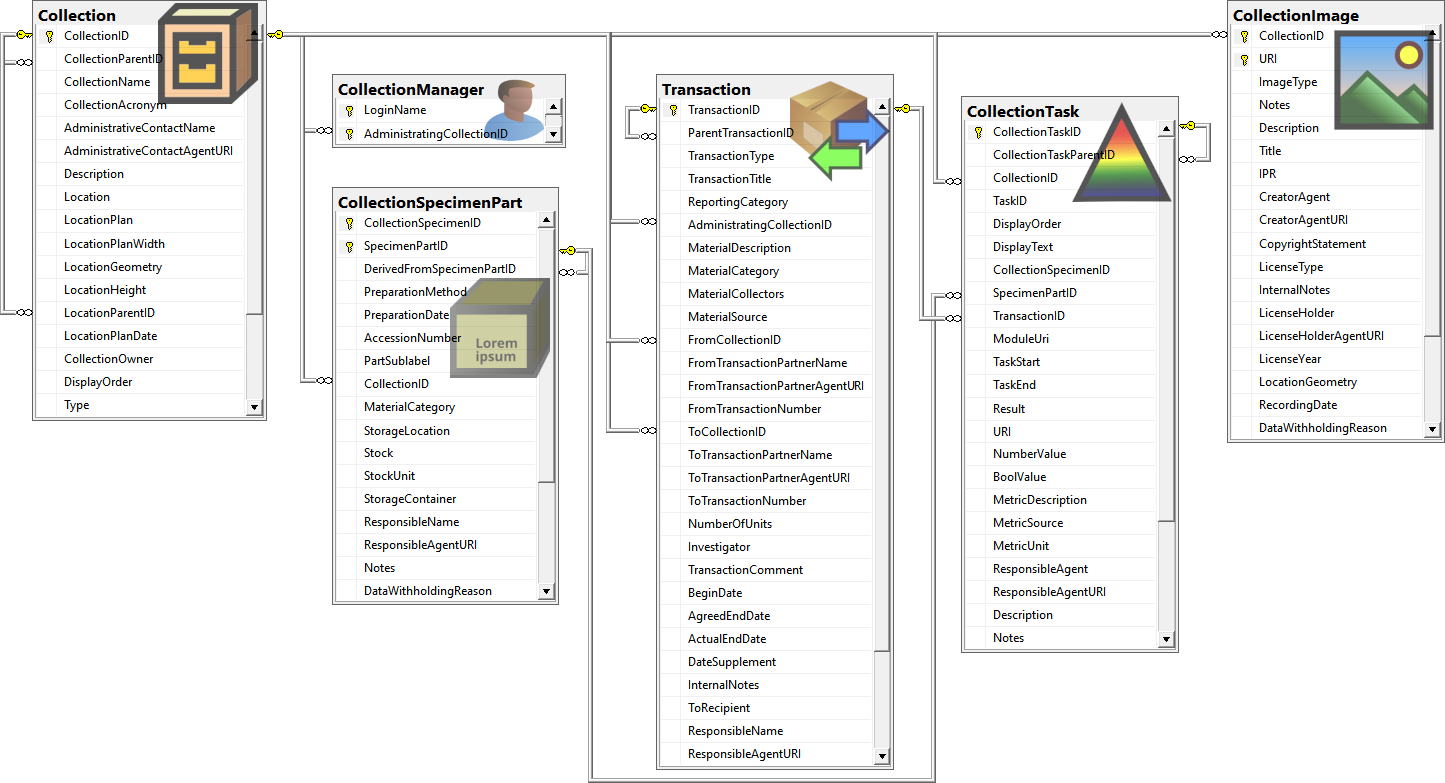
Diversity Collection
Project Tables
The image below shows the tables related to the project.
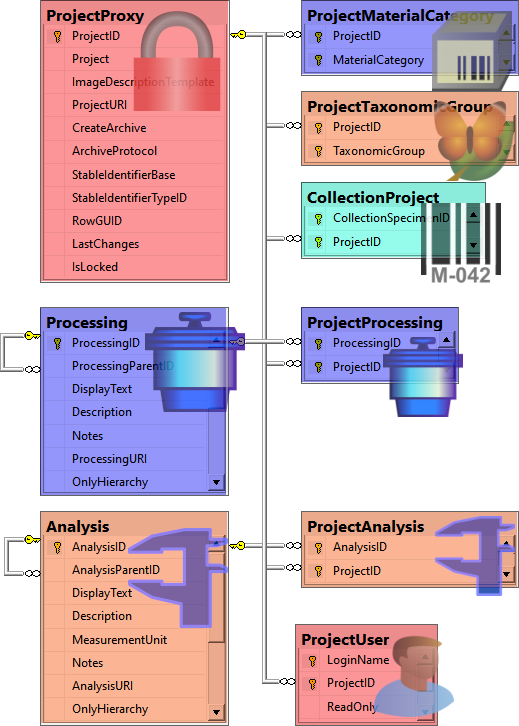
Diversity Collection
Analysis Tables
The image below shows the tables related to the  analysis and the
analysis and the  methods used for the analysis and charaterized by
methods used for the analysis and charaterized by  parameters. The main focus of object data are the
parameters. The main focus of object data are the  identification units (= organisms) with an optional restriction to a certain
identification units (= organisms) with an optional restriction to a certain  specimen part.
specimen part.
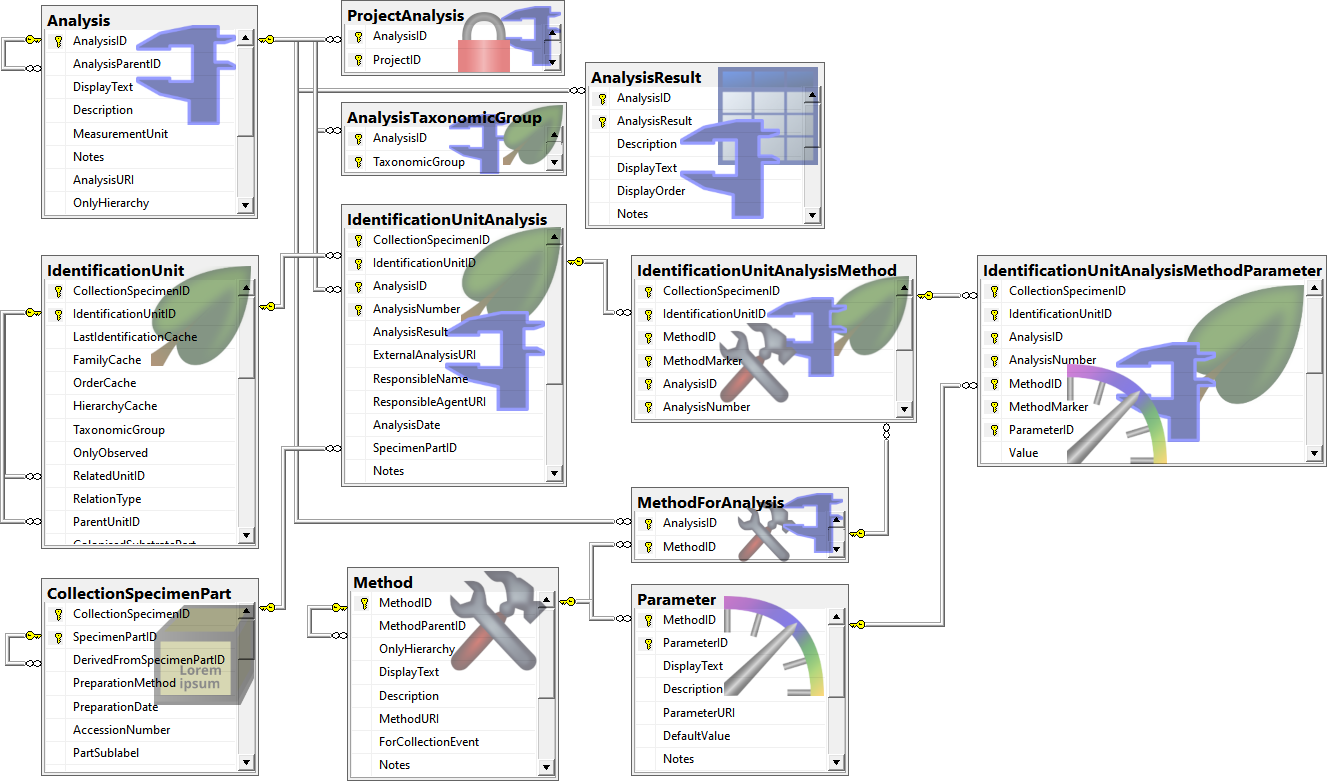
Links to the details of the tables
Diversity Collection
Processing Tables
The image below shows the tables related to the Processing.
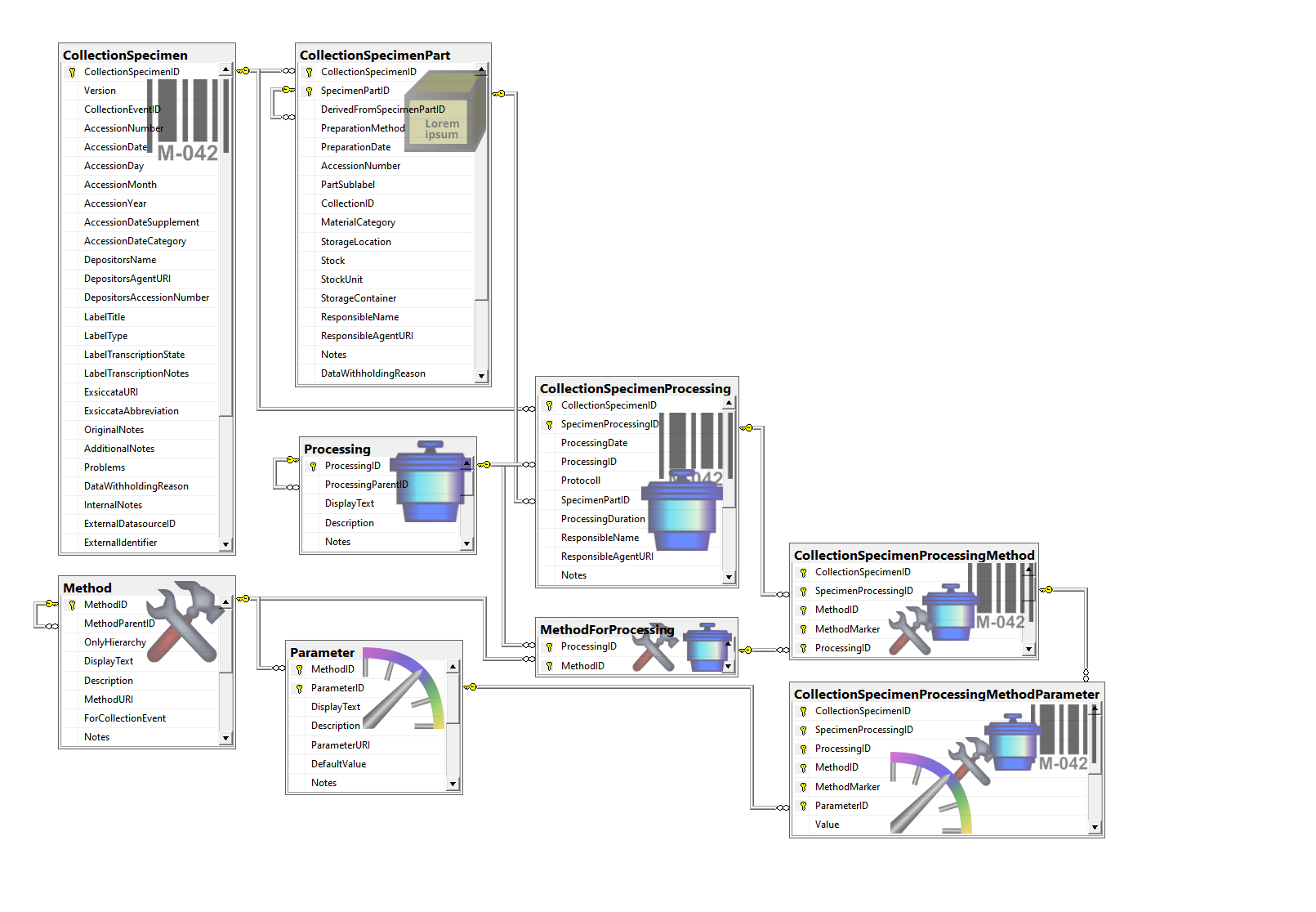
Diversity Collection
Transaction Tables
The image below shows the tables related to the Transaction.
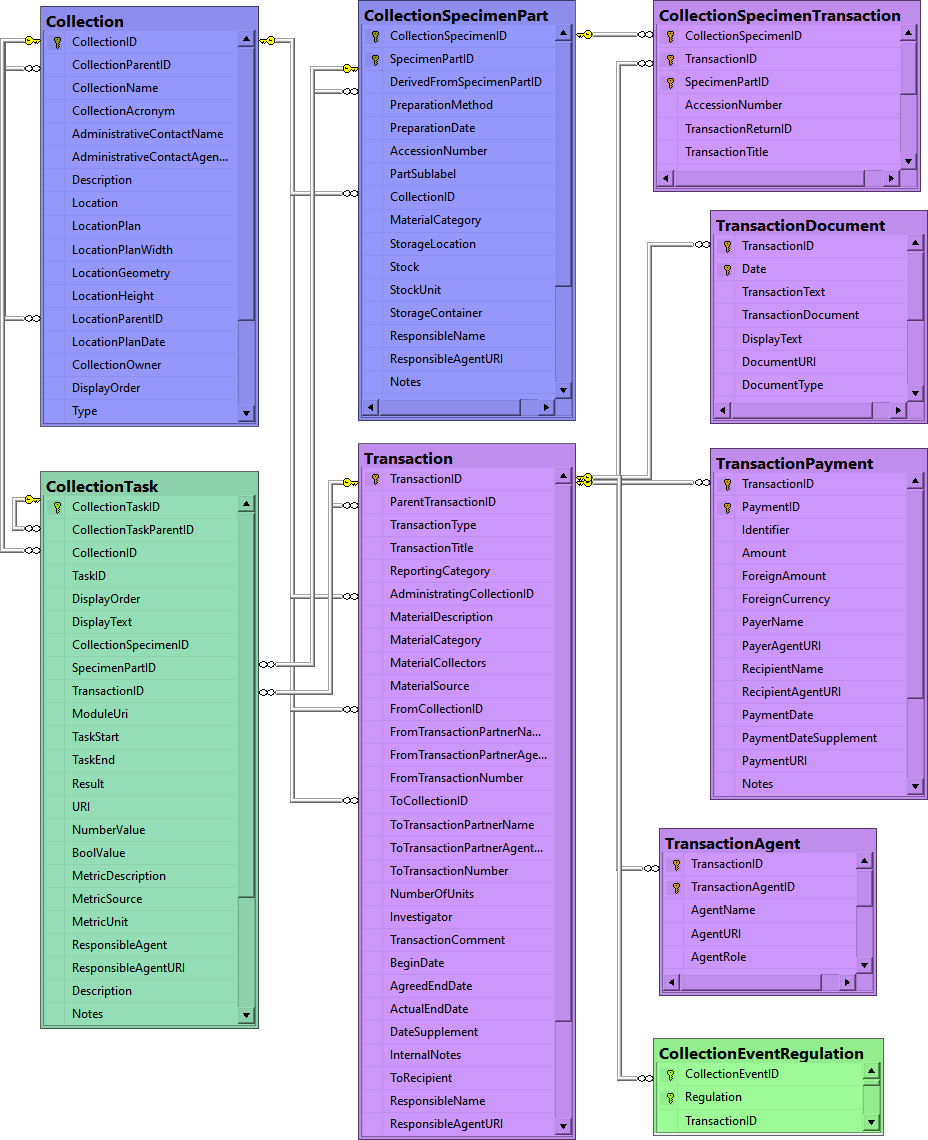
Diversity Collection
Task Tables
The image below shows the tables related to the task.
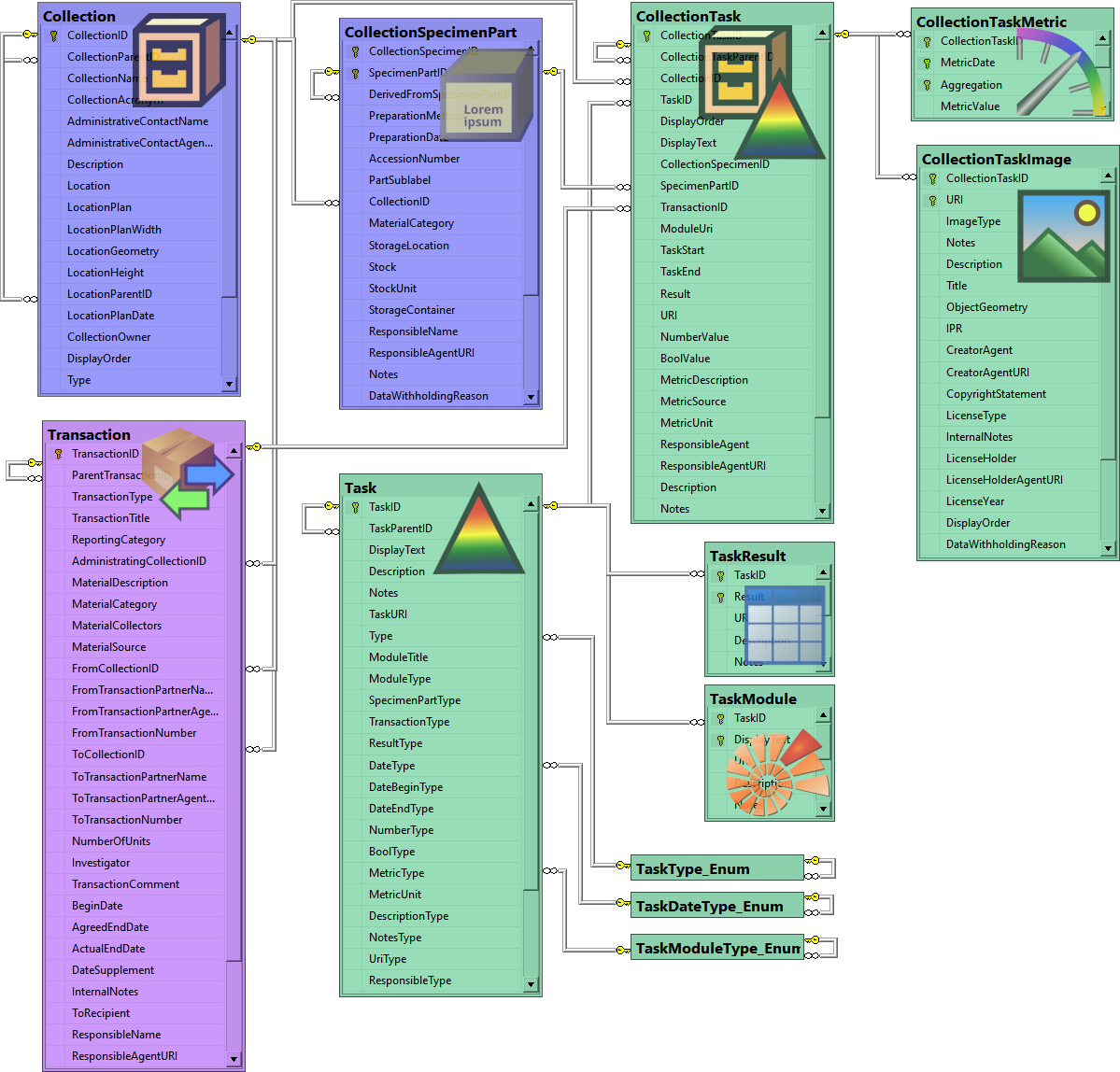
Diversity Collection
Entity Tables
The image below shows the tables related to the Entity.
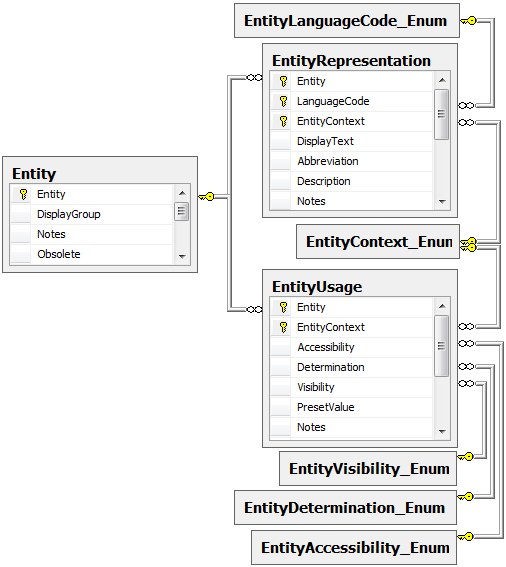
Diversity Collection
Mapping
According to Specifications
Subsections of Mapping
Diversity Collection
Mapping
Diversity Collection
Mapping
Diversity Collection
Mapping
Darwin Core
copilot
1. Versuch
| subject_id |
predicate_id |
object_id |
subject_label |
object_label |
mapping_justification |
| CollectionEvent.CollectionDate |
skos:exactMatch |
dwc:eventDate |
Collection Date |
Event Date |
Identical semantic meaning |
| CollectionEvent.StartDate |
skos:closeMatch |
dwc:eventDate |
Start Date |
Event Date |
Partial overlap |
| CollectionEvent.EndDate |
skos:closeMatch |
dwc:eventDate |
End Date |
Event Date |
Partial overlap |
| CollectionEvent.Method |
skos:exactMatch |
dwc:samplingProtocol |
Collection Method |
Sampling Protocol |
Equivalent terminology |
| CollectionEvent.LocationID |
skos:exactMatch |
dwc:locationID |
Location ID |
Location ID |
Identical semantic meaning |
| CollectionEvent.Latitude |
skos:exactMatch |
dwc:decimalLatitude |
Latitude |
Decimal Latitude |
Identical semantic meaning |
| CollectionEvent.Longitude |
skos:exactMatch |
dwc:decimalLongitude |
Longitude |
Decimal Longitude |
Identical semantic meaning |
| CollectionEvent.Elevation |
skos:closeMatch |
dwc:verbatimElevation |
Elevation |
Verbatim Elevation |
Similar but not standardized |
| CollectionEvent.Habitat |
skos:exactMatch |
dwc:habitat |
Habitat |
Habitat |
Identical semantic meaning |
| CollectionEvent.Collector |
skos:exactMatch |
dwc:recordedBy |
Collector |
Recorded By |
Equivalent terminology |
| CollectionEvent.Remarks |
skos:closeMatch |
dwc:eventRemarks |
Remarks |
Event Remarks |
Partial overlap |
2. Versuch
please create a sssom mapping to darwincore (https://dwc.tdwg.org) for the database table CollectionEvent as defined below:
### Table **CollectionEvent**
The event where and when the specimen were collected
| Column | Data type | Description | Nullable | Relation |
| --- | :-: | --- | :-: | --- |
| __CollectionEventID__| int| Unique ID for the table CollectionEvent (= primary key)| NO| -|
| Version| int| The version of the data set. Automatically set by the system.Default value: (1)| NO| -|
| SeriesID| int| The ID of the related expedition. Relates to the PK of the table CollectionExpedition (foreign key). | YES| Refers to table CollectionEventSeries|
| CollectorsEventNumber| nvarchar (50)| Number assigned to a collection event by the collector (= 'field number')| YES| -|
| CollectionDate| datetime| The cached date of the collection event calulated from the entries in CollectionDay, -Month and -Year.| YES| -|
| CollectionDay| tinyint| The day of the date of the event or when the collection event started| YES| -|
| CollectionMonth| tinyint| The month of the date of the event or when the collection event started| YES| -|
| CollectionYear| smallint| The year of the date of the event or when the collection event started| YES| -|
| CollectionEndDay| tinyint| The day of the date of the event or when the collection event ended| YES| -|
| CollectionEndMonth| tinyint| The month of the date of the event or when the collection event ended| YES| -|
| CollectionEndYear| smallint| The year of the date of the event or when the collection event ended| YES| -|
| CollectionDateSupplement| nvarchar (100)| Verbal or additional collection date information, e.g. 'end of summer 1985', 'first quarter', '1888-1892'. The end date, if the collection event comprises a period. The time of the event, if necessary.| YES| -|
| CollectionDateCategory| nvarchar (50)| Category of the date of the identification e.g. "system", "estimated" (= foreign key, see in table CollEventDateCategory_Enum)| YES| Refers to table CollEventDateCategory_Enum|
| CollectionTime| varchar (50)| The time of the event or when the collection event started| YES| -|
| CollectionTimeSpan| varchar (50)| The time span e.g. in seconds of the collection event| YES| -|
| LocalityDescription| nvarchar (MAX)| Locality description of the locality exactly as written on the original label (i.e. without corrections during data entry)| YES| -|
| LocalityVerbatim| nvarchar (MAX)| Locality as given in historical context, documents and labels| YES| -|
| HabitatDescription| nvarchar (MAX)| Geo-ecological description of the locality exactly as written on the original label (i.e. without corrections during data entry)| YES| -|
| ReferenceTitle| nvarchar (255)| The title of the publication where the collection event was published. Note that this is only a cached value where ReferenceURI is present| YES| -|
| ReferenceURI| varchar (255)| URI (e.g. LSID) of the source publication where the collection event is published, may e.g. refer to the module DiversityReferences| YES| -|
| ReferenceDetails| nvarchar (50)| The exact location within the reference, e.g. pages, plates| YES| -|
| CollectingMethod| nvarchar (MAX)| Description of the method used for collecting the samples, e.g. traps, moist chambers, drag net| YES| -|
| Notes| nvarchar (MAX)| Notes on the collection event| YES| -|
| CountryCache| nvarchar (50)| The country where the collection event took place. Cached value derived from an entry in CollectionEventLocalisation| YES| -|
| DataWithholdingReason| nvarchar (255)| If the data set is withhold, the reason for withholding the data, otherwise null| YES| -|
| DataWithholdingReasonDate| nvarchar (50)| The reason for withholding the collection date| YES| -|
| LogCreatedWhen| datetime| Point in time when this data set was createdDefault value: getdate()| YES| -|
| LogCreatedBy| nvarchar (50)| Name of the creator of this data setDefault value: suser_sname()| YES| -|
| LogUpdatedWhen| datetime| Point in time when this data set was updated lastDefault value: getdate()| YES| -|
| LogUpdatedBy| nvarchar (50)| Name of the person to update this data set lastDefault value: suser_sname()| YES| -|
SSSOM Mapping: DiversityCollection.CollectionEvent → Darwin Core
Mapping Table
| subject_id |
predicate_id |
object_id |
subject_label |
object_label |
mapping_justification |
| CollectionEventID |
skos:exactMatch |
dwc:eventID |
Collection Event ID |
Event ID |
Unique identifier for the event |
| CollectorsEventNumber |
skos:closeMatch |
dwc:fieldNumber |
Collector’s Event Number |
Field Number |
Collector-assigned identifier |
| CollectionDate |
skos:exactMatch |
dwc:eventDate |
Collection Date |
Event Date |
Standardized collection date |
| CollectionDay |
skos:relatedMatch |
dwc:eventDate |
Collection Day |
Event Date |
Partial date component |
| CollectionMonth |
skos:relatedMatch |
dwc:eventDate |
Collection Month |
Event Date |
Partial date component |
| CollectionYear |
skos:relatedMatch |
dwc:eventDate |
Collection Year |
Event Date |
Partial date component |
| CollectionEndDay |
skos:relatedMatch |
dwc:endDayOfYear |
Collection End Day |
End Day of Year |
End date component |
| CollectionEndMonth |
skos:relatedMatch |
dwc:endMonth |
Collection End Month |
End Month |
End date component |
| CollectionEndYear |
skos:relatedMatch |
dwc:endYear |
Collection End Year |
End Year |
End date component |
| CollectionDateSupplement |
skos:closeMatch |
dwc:eventRemarks |
Collection Date Supplement |
Event Remarks |
Verbal or uncertain date info |
| CollectionDateCategory |
skos:relatedMatch |
dwc:eventRemarks |
Collection Date Category |
Event Remarks |
Qualifier for date certainty |
| CollectionTime |
skos:closeMatch |
dwc:eventTime |
Collection Time |
Event Time |
Time of collection |
| CollectionTimeSpan |
skos:relatedMatch |
dwc:eventRemarks |
Collection Time Span |
Event Remarks |
Duration of collection |
| LocalityDescription |
skos:exactMatch |
dwc:verbatimLocality |
Locality Description |
Verbatim Locality |
Label-based locality description |
| LocalityVerbatim |
skos:closeMatch |
dwc:verbatimLocality |
Locality Verbatim |
Verbatim Locality |
Historical locality phrasing |
| HabitatDescription |
skos:exactMatch |
dwc:habitat |
Habitat Description |
Habitat |
Ecological context of locality |
| ReferenceTitle |
skos:relatedMatch |
dwc:associatedReferences |
Reference Title |
Associated References |
Title of related publication |
| ReferenceURI |
skos:exactMatch |
dwc:associatedReferences |
Reference URI |
Associated References |
URI of related publication |
| ReferenceDetails |
skos:relatedMatch |
dwc:associatedReferences |
Reference Details |
Associated References |
Specific location in reference |
| CollectingMethod |
skos:exactMatch |
dwc:samplingProtocol |
Collecting Method |
Sampling Protocol |
Method used for collection |
| Notes |
skos:closeMatch |
dwc:eventRemarks |
Notes |
Event Remarks |
General notes on event |
| CountryCache |
skos:exactMatch |
dwc:country |
Country |
Country |
Country of collection |
| DataWithholdingReason |
skos:relatedMatch |
dwc:dataGeneralizations |
Data Withholding Reason |
Data Generalizations |
Reason for data suppression |
| DataWithholdingReasonDate |
skos:relatedMatch |
dwc:dataGeneralizations |
Data Withholding Reason Date |
Data Generalizations |
Reason for date suppression |
| LogCreatedWhen |
skos:relatedMatch |
dwc:modified |
Log Created When |
Modified |
Metadata timestamp |
| LogCreatedBy |
skos:relatedMatch |
dwc:recordedBy |
Log Created By |
Recorded By |
Creator of record |
| LogUpdatedWhen |
skos:relatedMatch |
dwc:modified |
Log Updated When |
Modified |
Last update timestamp |
| LogUpdatedBy |
skos:relatedMatch |
dwc:recordedBy |
Log Updated By |
Recorded By |
Last updater of record |
- Mapping Creator: Microsoft Copilot
- Mapping Date: 2025-09-17
- License: CC-BY 4.0
- Subject Source: DiversityCollection (CollectionEvent Table)
- Object Source: Darwin Core Standard (https://dwc.tdwg.org)
3. Versuch
please create a sssom mapping to darwincore (https://dwc.tdwg.org) including all sssom columns for the database table Analysis as defined below:
### Table **Analysis**
Analysis types used within the database
| Column | Data type | Description | Nullable | Relation |
| --- | :-: | --- | :-: | --- |
| __AnalysisID__| int| ID of the analysis (primary key)| NO| -|
| AnalysisParentID| int| Analysis ID of the parent analysis, if it belongs to a certain type documented in this table| YES| Refers to table Analysis|
| DisplayText| nvarchar (50)| Name of the analysis as e.g. shown in user interface| YES| -|
| Description| nvarchar (MAX)| Description of the analysis| YES| -|
| MeasurementUnit| nvarchar (50)| The measurement unit used for the analysis, e.g. mm, µmol, kg| YES| -|
| Notes| nvarchar (MAX)| Notes on this analysis| YES| -|
| AnalysisURI| varchar (255)| URI referring to an external documentation of the analysis| YES| -|
| OnlyHierarchy| bit| If the entry is only used for the hierarchical arrangement of the entriesDefault value: (0)| YES| -|
| LogCreatedWhen| datetime| Point in time when this data set was createdDefault value: getdate()| YES| -|
| LogCreatedBy| nvarchar (50)| Name of the creator of this data setDefault value: suser_sname()| YES| -|
| LogUpdatedWhen| datetime| Point in time when this data set was updated lastDefault value: getdate()| YES| -|
| LogUpdatedBy| nvarchar (50)| Name of the person to update this data set lastDefault value: suser_sname()| YES| -|
| RowGUID| uniqueidentifier| -Default value: newsequentialid()| NO| -|
Result in main page
Gemini
1. Versuch
title: “CollectionEvent to Darwin Core SSSOM Mapping”
description: “A standard mapping from the Diversity Workbench CollectionEvent table to Darwin Core terms.”
date: 2023-10-27T10:00:00+02:00
draft: false
This document provides a mapping of columns from a typical Diversity Workbench CollectionEvent table to standard Darwin Core terms, following the SSSOM (Simple Standard for Sharing Ontology Mappings) schema.
| subject_id |
subject_label |
predicate_id |
object_id |
object_label |
mapping_justification |
comment |
CollectionEventID |
Collection Event ID |
skos:exactMatch |
dwc:eventID |
Event ID |
semap:LexicalMatch |
The unique identifier for the collection event. |
CollectorsName |
Collectors Name |
skos:exactMatch |
dwc:recordedBy |
Recorded By |
semap:LexicalMatch |
The name(s) of the person(s) who collected the item. |
VerbatimEventDate |
Verbatim Event Date |
skos:exactMatch |
dwc:verbatimEventDate |
Verbatim Event Date |
semap:LexicalMatch |
The verbatim original date. |
EventDate |
Event Date |
skos:exactMatch |
dwc:eventDate |
Event Date |
semap:DerivedMatch |
The standardized date of the event. |
Locality |
Locality |
skos:exactMatch |
dwc:locality |
Locality |
semap:LexicalMatch |
The specific description of the place. |
Latitude |
Latitude |
skos:exactMatch |
dwc:decimalLatitude |
Decimal Latitude |
semap:LexicalMatch |
The latitude in decimal degrees. |
Longitude |
Longitude |
skos:exactMatch |
dwc:decimalLongitude |
Decimal Longitude |
semap:LexicalMatch |
The longitude in decimal degrees. |
GeodeticDatum |
Geodetic Datum |
skos:exactMatch |
dwc:geodeticDatum |
Geodetic Datum |
semap:LexicalMatch |
The geodetic datum for the coordinates. |
Altitude |
Altitude |
skos:closeMatch |
dwc:verbatimElevation |
Verbatim Elevation |
semap:LexicalMatch |
The verbatim original elevation. Use dwc:minimumElevationInMeters and dwc:maximumElevationInMeters for a range. |
CoordinateUncertainty |
Coordinate Uncertainty |
skos:exactMatch |
dwc:coordinateUncertaintyInMeters |
Coordinate Uncertainty In Meters |
semap:LexicalMatch |
The horizontal distance in meters. |
Habitat |
Habitat |
skos:exactMatch |
dwc:habitat |
Habitat |
semap:LexicalMatch |
A description of the habitat. |
CollectingMethod |
Collecting Method |
skos:exactMatch |
dwc:samplingProtocol |
Sampling Protocol |
semap:LexicalMatch |
The method used to collect the specimen. |
Notes |
Notes |
skos:exactMatch |
dwc:eventRemarks |
Event Remarks |
semap:LexicalMatch |
General notes or remarks about the event. |
CollectionID |
Collection ID |
skos:closeMatch |
dwc:occurrenceID |
Occurrence ID |
semap:DerivedMatch |
The original ID from the source database, which might relate to the occurrence record. |
2. Versuch
erzeuge ein sssom mapping nach darwincore (https://dwc.tdwg.org) der datenbank tabelle CollectionEvent die hier definiert ist: https://www.diversityworkbench.de/manual/dwb_latest/modules/diversitycollection/internals_dc/database_dc/database_structure_dc/database_tables_dc/index.html#table-collectionevent. Halte Dich strikt an die Definition der tabelle CollectionEvent. Füge keine Spalten hinzu und lasse keine weg.
| subject_id (Spalte in CollectionEvent) |
predicate_id (Beziehung) |
object_id (Darwin Core Begriff) |
| CollectionEventID |
skos:closeMatch |
dwc:eventID |
| CollectionID |
skos:closeMatch |
dwc:collectionID |
| Collector |
skos:closeMatch |
dwc:recordedBy |
| LocalityDescription |
skos:closeMatch |
dwc:locality |
| Altitude |
skos:closeMatch |
dwc:minimumElevationInMeters |
| Altitude |
skos:closeMatch |
dwc:maximumElevationInMeters |
| AltitudeUnit |
skos:closeMatch |
dwc:verbatimElevation |
| Longitude |
skos:closeMatch |
dwc:decimalLongitude |
| Latitude |
skos:closeMatch |
dwc:decimalLatitude |
| GeoRefMethod |
skos:closeMatch |
dwc:georeferenceProtocol |
| HabitatDescription |
skos:closeMatch |
dwc:habitat |
| DateStart |
skos:closeMatch |
dwc:eventDate |
| DateEnd |
skos:closeMatch |
dwc:eventDate |
| CollectionTime |
skos:closeMatch |
dwc:eventTime |
| Notes |
skos:closeMatch |
dwc:eventRemarks |
| Number |
skos:closeMatch |
dwc:recordNumber |
| LocationID |
skos:closeMatch |
dwc:locationID |
| AccessionDate |
skos:closeMatch |
dwc:dateCollected |
| AccessionNumber |
skos:closeMatch |
dwc:catalogNumber |
| SeriesNumber |
skos:closeMatch |
dwc:collectionNumber |
| Author |
skos:closeMatch |
dwc:recordedBy |
| Version |
skos:closeMatch |
dwc:dataGeneralizations |
please create a sssom mapping to darwincore (https://dwc.tdwg.org) for the database table CollectionEvent as defined below:
### Table **CollectionEvent**
The event where and when the specimen were collected
| Column | Data type | Description | Nullable | Relation |
| --- | :-: | --- | :-: | --- |
| __CollectionEventID__| int| Unique ID for the table CollectionEvent (= primary key)| NO| -|
| Version| int| The version of the data set. Automatically set by the system.Default value: (1)| NO| -|
| SeriesID| int| The ID of the related expedition. Relates to the PK of the table CollectionExpedition (foreign key). | YES| Refers to table CollectionEventSeries|
| CollectorsEventNumber| nvarchar (50)| Number assigned to a collection event by the collector (= 'field number')| YES| -|
| CollectionDate| datetime| The cached date of the collection event calulated from the entries in CollectionDay, -Month and -Year.| YES| -|
| CollectionDay| tinyint| The day of the date of the event or when the collection event started| YES| -|
| CollectionMonth| tinyint| The month of the date of the event or when the collection event started| YES| -|
| CollectionYear| smallint| The year of the date of the event or when the collection event started| YES| -|
| CollectionEndDay| tinyint| The day of the date of the event or when the collection event ended| YES| -|
| CollectionEndMonth| tinyint| The month of the date of the event or when the collection event ended| YES| -|
| CollectionEndYear| smallint| The year of the date of the event or when the collection event ended| YES| -|
| CollectionDateSupplement| nvarchar (100)| Verbal or additional collection date information, e.g. 'end of summer 1985', 'first quarter', '1888-1892'. The end date, if the collection event comprises a period. The time of the event, if necessary.| YES| -|
| CollectionDateCategory| nvarchar (50)| Category of the date of the identification e.g. "system", "estimated" (= foreign key, see in table CollEventDateCategory_Enum)| YES| Refers to table CollEventDateCategory_Enum|
| CollectionTime| varchar (50)| The time of the event or when the collection event started| YES| -|
| CollectionTimeSpan| varchar (50)| The time span e.g. in seconds of the collection event| YES| -|
| LocalityDescription| nvarchar (MAX)| Locality description of the locality exactly as written on the original label (i.e. without corrections during data entry)| YES| -|
| LocalityVerbatim| nvarchar (MAX)| Locality as given in historical context, documents and labels| YES| -|
| HabitatDescription| nvarchar (MAX)| Geo-ecological description of the locality exactly as written on the original label (i.e. without corrections during data entry)| YES| -|
| ReferenceTitle| nvarchar (255)| The title of the publication where the collection event was published. Note that this is only a cached value where ReferenceURI is present| YES| -|
| ReferenceURI| varchar (255)| URI (e.g. LSID) of the source publication where the collection event is published, may e.g. refer to the module DiversityReferences| YES| -|
| ReferenceDetails| nvarchar (50)| The exact location within the reference, e.g. pages, plates| YES| -|
| CollectingMethod| nvarchar (MAX)| Description of the method used for collecting the samples, e.g. traps, moist chambers, drag net| YES| -|
| Notes| nvarchar (MAX)| Notes on the collection event| YES| -|
| CountryCache| nvarchar (50)| The country where the collection event took place. Cached value derived from an entry in CollectionEventLocalisation| YES| -|
| DataWithholdingReason| nvarchar (255)| If the data set is withhold, the reason for withholding the data, otherwise null| YES| -|
| DataWithholdingReasonDate| nvarchar (50)| The reason for withholding the collection date| YES| -|
| LogCreatedWhen| datetime| Point in time when this data set was createdDefault value: getdate()| YES| -|
| LogCreatedBy| nvarchar (50)| Name of the creator of this data setDefault value: suser_sname()| YES| -|
| LogUpdatedWhen| datetime| Point in time when this data set was updated lastDefault value: getdate()| YES| -|
| LogUpdatedBy| nvarchar (50)| Name of the person to update this data set lastDefault value: suser_sname()| YES| -|
The mapping should contain the sssom columns subject_id, subject_label, predicate_id, object_id, object_label, mapping_justification, comment.
3. Versuch mit Gemini
Here is the SSSOM mapping for the CollectionEvent table to Darwin Core, based on the provided definition.
| subject_id |
subject_label |
predicate_id |
object_id |
object_label |
mapping_justification |
comment |
| CollectionEventID |
CollectionEventID |
skos:closeMatch |
dwc:eventID |
eventID |
This column serves as a unique identifier for the collection event. |
Unique identifier for a collection event. |
| Version |
Version |
skos:closeMatch |
dwc:datasetID |
datasetID |
The dataset version can be mapped to a unique dataset identifier. |
The version of the data set. |
| SeriesID |
SeriesID |
skos:closeMatch |
dwc:eventID |
eventID |
This ID relates to a series of collection events, which could be represented as an event ID. |
Relates to the collection expedition or series of events. |
| CollectorsEventNumber |
CollectorsEventNumber |
skos:closeMatch |
dwc:recordNumber |
recordNumber |
This field number is the collector’s identifier for a specific collection event. |
Number assigned to a collection event by the collector. |
| CollectionDate |
CollectionDate |
skos:closeMatch |
dwc:eventDate |
eventDate |
The cached date of the collection event is a direct match for eventDate. |
The cached date of the collection event. |
| CollectionDay |
CollectionDay |
skos:closeMatch |
dwc:day |
day |
Direct mapping to the Darwin Core day term. |
The day of the date of the event. |
| CollectionMonth |
CollectionMonth |
skos:closeMatch |
dwc:month |
month |
Direct mapping to the Darwin Core month term. |
The month of the date of the event. |
| CollectionYear |
CollectionYear |
skos:closeMatch |
dwc:year |
year |
Direct mapping to the Darwin Core year term. |
The year of the date of the event. |
| CollectionEndDay |
CollectionEndDay |
skos:closeMatch |
dwc:endDayOfYear |
endDayOfYear |
Represents the end day of a multi-day collection event. |
The day of the end of the collection event. |
| CollectionEndMonth |
CollectionEndMonth |
skos:closeMatch |
dwc:endDayOfYear |
endDayOfYear |
Represents the end month of a multi-month collection event. |
The month of the end of the collection event. |
| CollectionEndYear |
CollectionEndYear |
skos:closeMatch |
dwc:year |
year |
The end year of a multi-year collection event. |
The year of the end of the collection event. |
| CollectionDateSupplement |
CollectionDateSupplement |
skos:closeMatch |
dwc:verbatimEventDate |
verbatimEventDate |
This column contains verbatim date information not suitable for structured fields. |
Verbal or additional collection date information. |
| CollectionDateCategory |
CollectionDateCategory |
skos:closeMatch |
dwc:eventRemarks |
eventRemarks |
This is a categorical note about the date and can be included in general remarks. |
Category of the date of the identification. |
| CollectionTime |
CollectionTime |
skos:closeMatch |
dwc:eventTime |
eventTime |
Direct mapping to the Darwin Core event time term. |
The time of the event. |
| CollectionTimeSpan |
CollectionTimeSpan |
skos:closeMatch |
dwc:duration |
duration |
Represents the duration of the event. |
The time span of the collection event. |
| LocalityDescription |
LocalityDescription |
skos:closeMatch |
dwc:locality |
locality |
Direct mapping to the Darwin Core locality term. |
Locality description on the original label. |
| LocalityVerbatim |
LocalityVerbatim |
skos:closeMatch |
dwc:verbatimLocality |
verbatimLocality |
This column explicitly holds verbatim locality information. |
Locality as given in historical context. |
| HabitatDescription |
HabitatDescription |
skos:closeMatch |
dwc:habitat |
habitat |
Direct mapping to the Darwin Core habitat term. |
Geo-ecological description of the locality. |
| ReferenceTitle |
ReferenceTitle |
skos:closeMatch |
dwc:bibliographicCitation |
bibliographicCitation |
The title of a publication is a core component of a bibliographic citation. |
The title of the publication where the collection event was published. |
| ReferenceURI |
ReferenceURI |
skos:closeMatch |
dwc:occurrenceID |
occurrenceID |
A URI referencing a publication can be used to uniquely identify an occurrence. |
URI of the source publication where the collection event is published. |
| ReferenceDetails |
ReferenceDetails |
skos:closeMatch |
dwc:bibliographicCitation |
bibliographicCitation |
Details like page numbers are part of a full bibliographic citation. |
The exact location within the reference. |
| CollectingMethod |
CollectingMethod |
skos:closeMatch |
dwc:samplingProtocol |
samplingProtocol |
This is a direct description of the method used for collecting. |
Description of the method used for collecting. |
| Notes |
Notes |
skos:closeMatch |
dwc:eventRemarks |
eventRemarks |
General notes about the event. |
Notes on the collection event. |
| CountryCache |
CountryCache |
skos:closeMatch |
dwc:country |
country |
Direct mapping to the Darwin Core country term. |
The country where the collection event took place. |
| DataWithholdingReason |
DataWithholdingReason |
skos:closeMatch |
dwc:dataGeneralizations |
dataGeneralizations |
The reason for withholding data relates to the generalization of the data to protect sensitive information. |
If the data set is withheld, the reason for withholding. |
| DataWithholdingReasonDate |
DataWithholdingReasonDate |
skos:closeMatch |
dwc:eventRemarks |
eventRemarks |
This is a remark about why the date is withheld. |
The reason for withholding the collection date. |
| LogCreatedWhen |
LogCreatedWhen |
skos:closeMatch |
dwc:dateIdentified |
dateIdentified |
The creation log can be mapped to the date of identification. |
Point in time when this data set was created. |
| LogCreatedBy |
LogCreatedBy |
skos:closeMatch |
dwc:recordedBy |
recordedBy |
The name of the creator can be mapped to the person who recorded the event. |
Name of the creator of this data set. |
| LogUpdatedWhen |
LogUpdatedWhen |
skos:closeMatch |
dwc:dateIdentified |
dateIdentified |
The log update time can be a proxy for a later date of identification. |
Point in time when this data set was updated last. |
| LogUpdatedBy |
LogUpdatedBy |
skos:closeMatch |
dwc:recordedBy |
recordedBy |
The name of the person who updated the data can be mapped to the recorder. |
Name of the person to update this data set last. |
Versuch mit Claude
🧬 SSSOM Mapping: CollectionEvent → Darwin Core
| subject_id |
subject_label |
predicate_id |
object_id |
object_label |
mapping_justification |
confidence |
comment |
| dc:CollectionEvent |
CollectionEvent |
skos:broadMatch |
dwc:Event |
Event |
CollectionEvent is a specialized form of Darwin Core Event representing specimen collection activities |
0.9 |
|
| dc:CollectionEventID |
CollectionEventID |
skos:exactMatch |
dwc:eventID |
eventID |
Primary key identifier for the collection event |
1.0 |
|
| dc:Version |
Version |
skos:noMatch |
|
none |
System versioning field - no direct Darwin Core equivalent |
1.0 |
|
| dc:SeriesID |
SeriesID |
skos:exactMatch |
dwc:parentEventID |
parentEventID |
References CollectionEventSeries which groups multiple events |
0.9 |
|
| dc:CollectorsEventNumber |
CollectorsEventNumber |
skos:exactMatch |
dwc:fieldNumber |
fieldNumber |
Field number assigned by collector to the collection event |
1.0 |
|
| dc:CollectionDate |
CollectionDate |
skos:exactMatch |
dwc:eventDate |
eventDate |
Cached date of the collection event |
1.0 |
|
| dc:CollectionDay |
CollectionDay |
skos:narrowMatch |
dwc:eventDate |
eventDate |
Day component of collection date |
0.8 |
|
| dc:CollectionMonth |
CollectionMonth |
skos:narrowMatch |
dwc:eventDate |
eventDate |
Month component of collection date |
0.8 |
|
| dc:CollectionYear |
CollectionYear |
skos:exactMatch |
dwc:year |
year |
Year component of collection date |
1.0 |
|
| dc:CollectionEndDay |
CollectionEndDay |
skos:narrowMatch |
dwc:eventDate |
eventDate |
End day for date ranges |
0.8 |
|
| dc:CollectionEndMonth |
CollectionEndMonth |
skos:narrowMatch |
dwc:eventDate |
eventDate |
End month for date ranges |
0.8 |
|
| dc:CollectionEndYear |
CollectionEndYear |
skos:narrowMatch |
dwc:eventDate |
eventDate |
End year for date ranges |
0.8 |
|
| dc:CollectionDateSupplement |
CollectionDateSupplement |
skos:narrowMatch |
dwc:eventDate |
eventDate |
Additional verbal date information |
0.7 |
|
| dc:CollectionDateCategory |
CollectionDateCategory |
skos:noMatch |
|
none |
Date category qualifier - no direct Darwin Core equivalent |
1.0 |
|
| dc:CollectionTime |
CollectionTime |
skos:exactMatch |
dwc:eventTime |
eventTime |
Time of collection event start |
1.0 |
|
| dc:CollectionTimeSpan |
CollectionTimeSpan |
skos:narrowMatch |
dwc:eventTime |
eventTime |
Duration of collection event |
0.8 |
|
| dc:LocalityDescription |
LocalityDescription |
skos:exactMatch |
dwc:locality |
locality |
Interpreted and potentially standardized locality description |
1.0 |
|
| dc:LocalityVerbatim |
LocalityVerbatim |
skos:exactMatch |
dwc:verbatimLocality |
verbatimLocality |
1:1 transcript of original locality information |
1.0 |
|
| dc:HabitatDescription |
HabitatDescription |
skos:exactMatch |
dwc:habitat |
habitat |
Geo-ecological habitat description |
1.0 |
|
| dc:ReferenceTitle |
ReferenceTitle |
skos:narrowMatch |
dcterms:bibliographicCitation |
bibliographicCitation |
Publication title where collection event was published |
0.8 |
|
| dc:ReferenceURI |
ReferenceURI |
skos:exactMatch |
dcterms:references |
references |
URI reference to source publication |
1.0 |
|
| dc:ReferenceDetails |
ReferenceDetails |
skos:narrowMatch |
dcterms:bibliographicCitation |
bibliographicCitation |
Specific location within reference (pages, plates) |
0.7 |
|
| dc:CollectingMethod |
CollectingMethod |
skos:exactMatch |
dwc:samplingProtocol |
samplingProtocol |
Method used for collecting samples |
1.0 |
|
| dc:Notes |
Notes |
skos:exactMatch |
dwc:fieldNotes |
fieldNotes |
General notes about the collection event |
0.9 |
|
| dc:CountryCache |
CountryCache |
skos:exactMatch |
dwc:country |
country |
Country where collection event took place |
1.0 |
|
| dc:DataWithholdingReason |
DataWithholdingReason |
skos:exactMatch |
dwc:informationWithheld |
informationWithheld |
Reason for withholding collection event data |
1.0 |
|
| dc:DataWithholdingReasonDate |
DataWithholdingReasonDate |
skos:narrowMatch |
dwc:informationWithheld |
informationWithheld |
Specific reason for withholding collection date |
0.8 |
|
| dc:LogCreatedWhen |
LogCreatedWhen |
skos:narrowMatch |
dcterms:created |
created |
Record creation timestamp |
0.8 |
|
| dc:LogCreatedBy |
LogCreatedBy |
skos:narrowMatch |
dcterms:creator |
creator |
Record creator |
0.7 |
|
| dc:LogUpdatedWhen |
LogUpdatedWhen |
skos:exactMatch |
dcterms:modified |
modified |
Record last modification timestamp |
1.0 |
|
| dc:LogUpdatedBy |
LogUpdatedBy |
skos:narrowMatch |
dcterms:contributor |
contributor |
Last person to update record |
0.7 |
|
| dc:RowGUID |
RowGUID |
skos:noMatch |
|
none |
System GUID field - no direct Darwin Core equivalent |
1.0 |
|
DiversityCollection
Model 2.6.47
(1 June, 2025)
|
|
| Authors |
M. Weiss, A. Grunz, G. Hagedorn, D. Triebel, S. Seifert |
| License |
CC BY-ND 3.0 |
| Suggested citation |
M. Weiss, A. Grunz, G. Hagedorn, D. Triebel & S. Seifert (2025). DiversityCollectioninformation model (version 2.6.47). http://www.diversityworkbench.net/Portal/DiversityCollectionModel_2.6.47 |
| Notes |
The model is implemented on MS SQL Server and the data types are also MS SQL Server specific |
Introduction:
DiversityCollection is part of the database framework
DiversityWorkbench. Within
this framework, the application DiversityCollection is confined to the
management of specimens in scientific collections. In this context, it is
designed to document any action concerning the collection, storage,
exchange, and treatment of specimens in a collection and is also
appropriate for storing observation data. DiversityCollection is
distinguished from other collection management systems by focusing on the
biological relationships between organisms collected together as one or more
specimens (e.g. host, parasite and hyperparasite, symbionts, etc.). Each
module within the Diversity Workbench is focused on a specific data
domain. DiversityCollection stores only data connected with the handling, provenience, identification and analysis
of collection specimens and observations. Data from other realms, such as
taxonomy, are handled in more detail in separate modules.
Data management sectors of the application
| Sector |
Description |
| Series |
An expedition, monitoring or similar that involves the collection event or observations at single spots |
| Event |
The location of an observation or the spot where a specimen was collected or observed during, for example, an expedition |
| Localisation |
Localisation of an observation or collection site |
| Site Property |
Properties of e.g. a collection site like the type of the habitat |
| Method |
Methods used for the observation or collection of specimen |
| Specimen |
Observed or collected specimen like those collected during a collection event |
| Collector |
The observers or collectors of specimen |
| Objects |
The objects like taxa or minerals observed or collected |
| Identification |
Identifications of the organisms, minerals etc. |
| Analysis |
Any analysis applied to characterise the objects |
| Analysis methods |
The methods that have been used to analyse the objects |
| Part |
The parts of a collected specimen that are stored in a collection, e.g. as herbarium sheets |
| Collection |
The collection where the collected objects are stored |
| Processing |
Processing applied to the parts of a specimen |
| Processing methods |
Methods that have been used for the processing |
| Transaction |
Actions like sending specimen as a loan to another collection |
| Project |
The projects that a specimen is a part of |
| Images |
Can be taken for the EventSeries, the CollectionEvent, the CollectionSpecimen as well as organisms and parts of a specimen |
| References |
The references related to a specimen, organism, identification or part of a specimen |
| Tasks |
The tasks related to a collection, e.g. IPM |
The following sections define the information model used in DiversityCollection. First the ER-diagrams are presented to give an overview over the relations between the entities/tables. Then the structure is separately documented for each table with textual information which attributes are stored in which column.
ER-Diagrams
The complete ER-Diagram is quite large thus the ER-Model is presented in separate, but overlapping diagrams.
Analysis

Collection
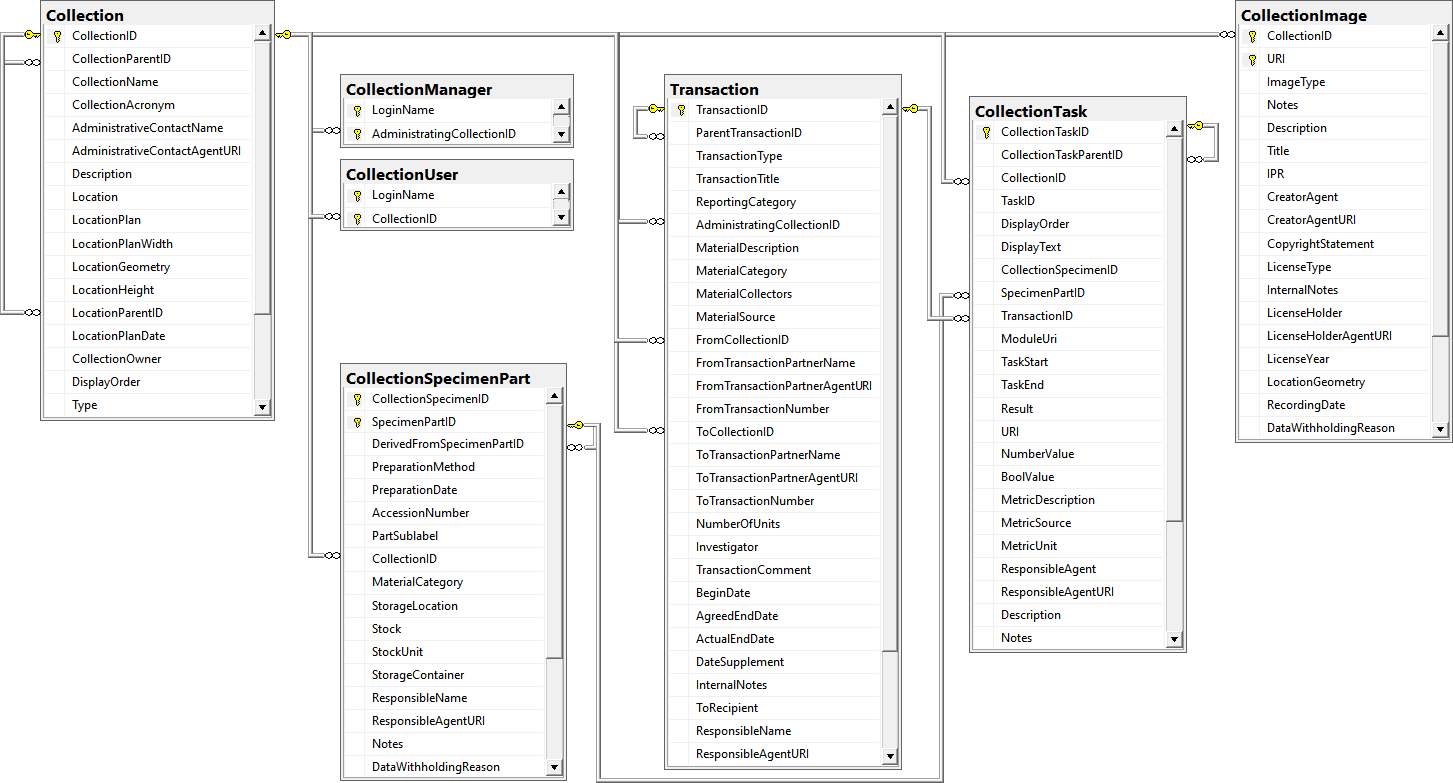
Event
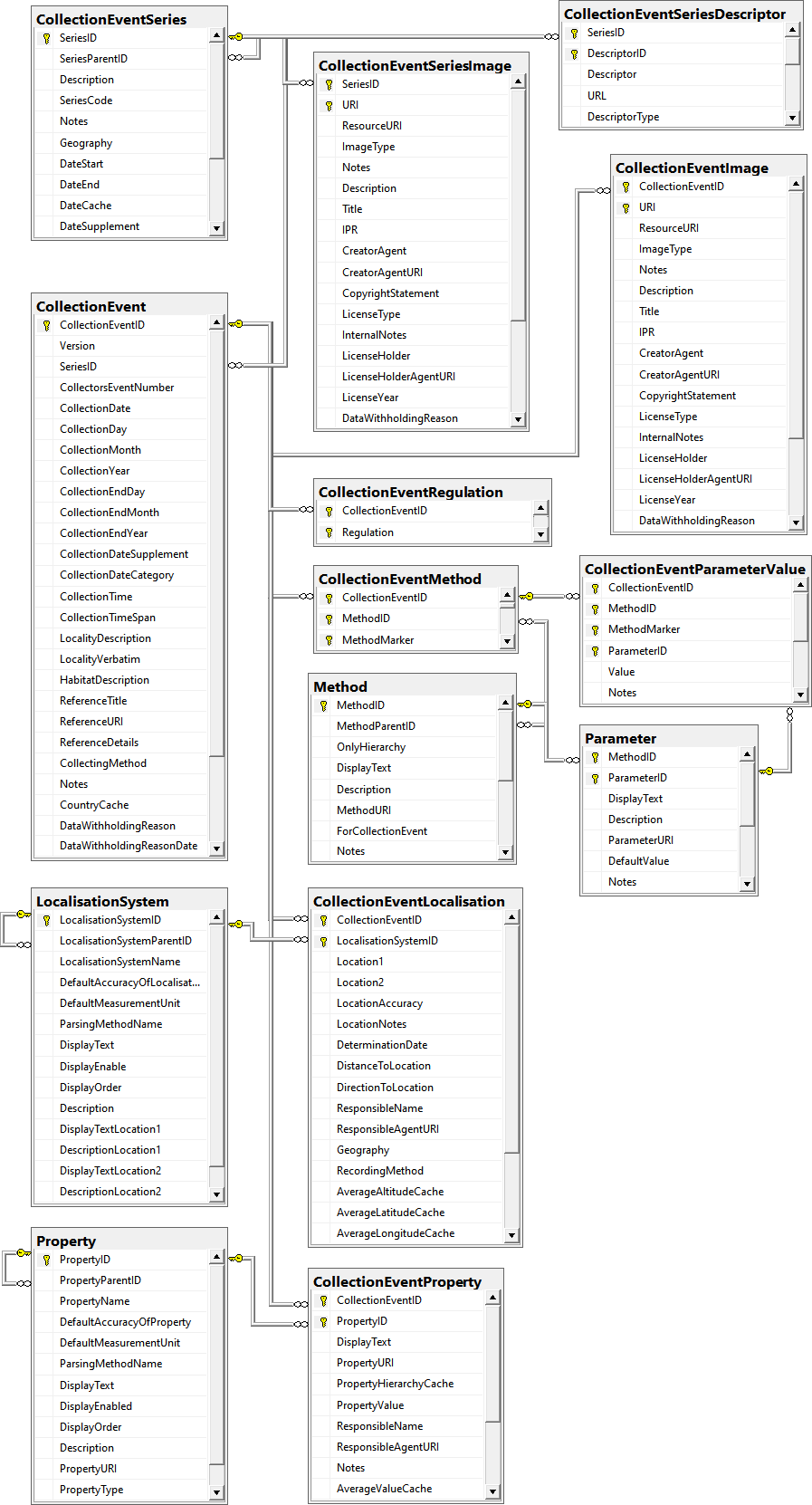
Method
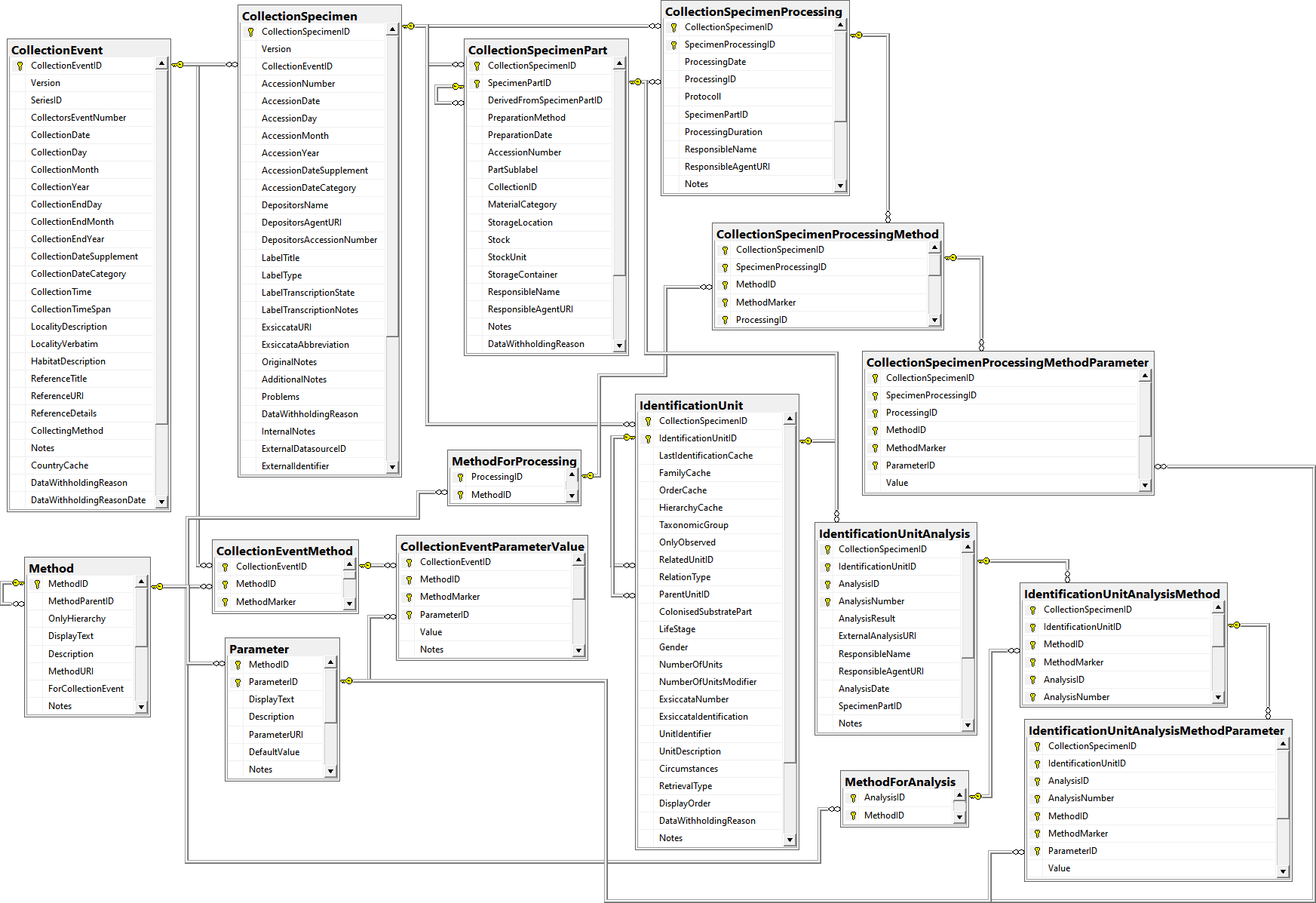
Part
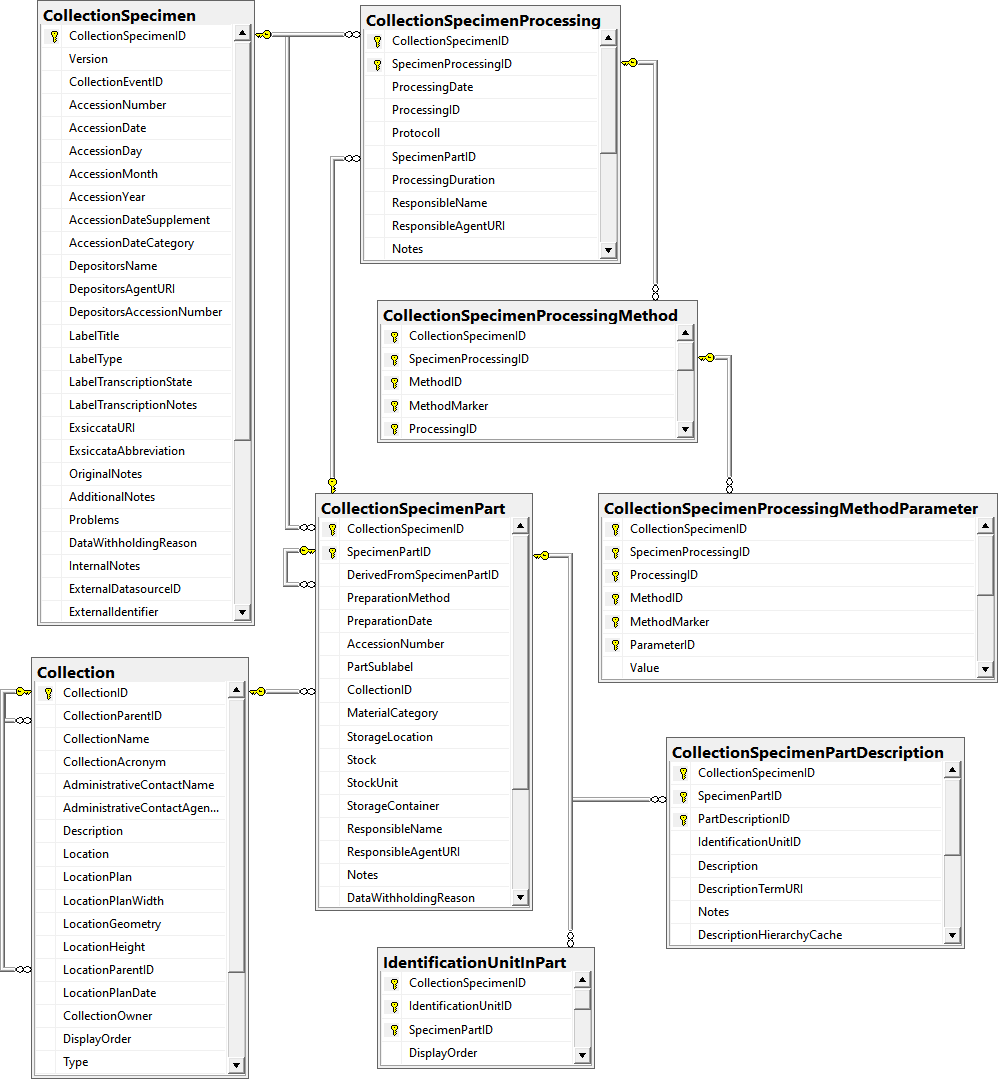
Processing
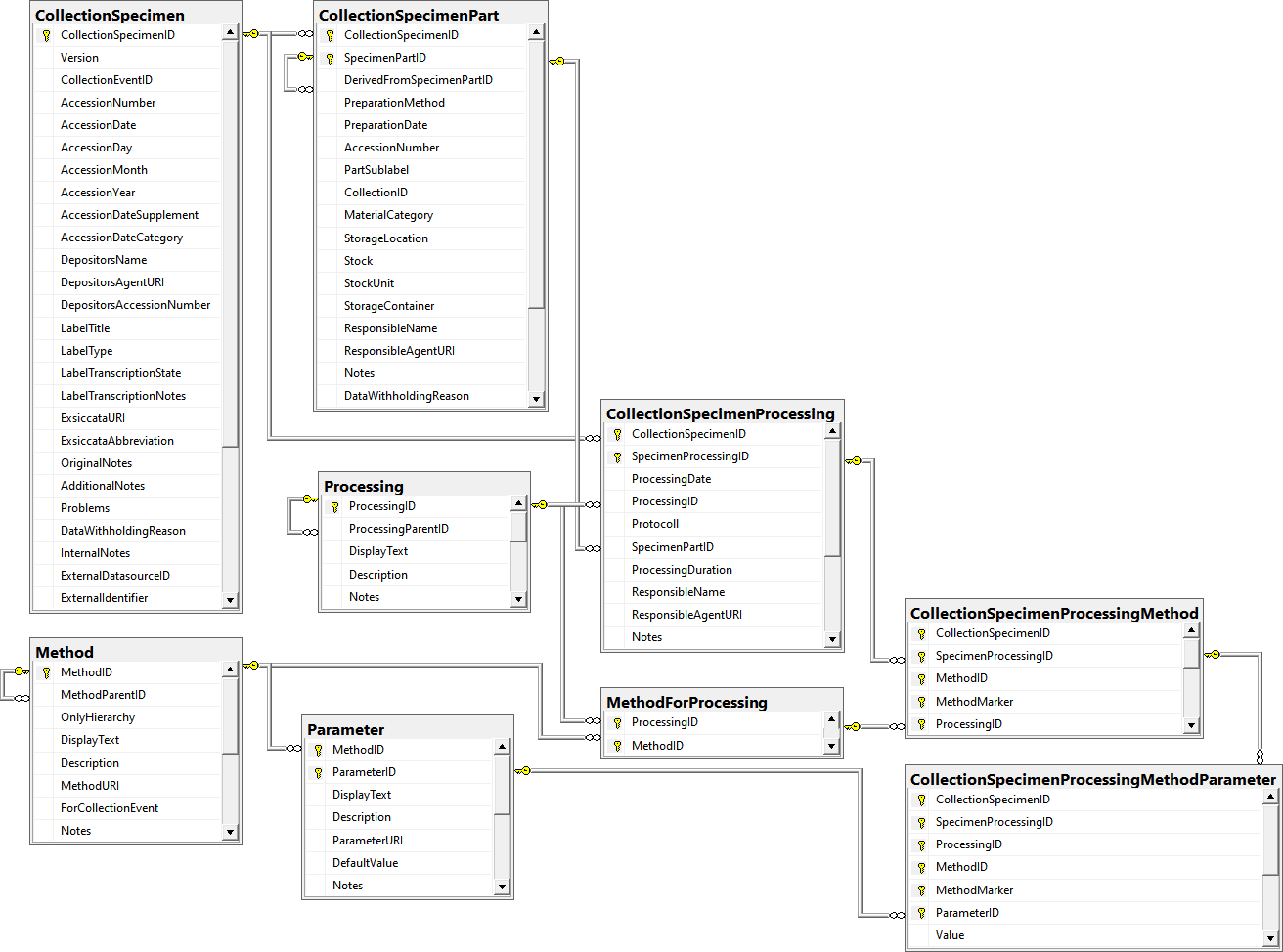
Project
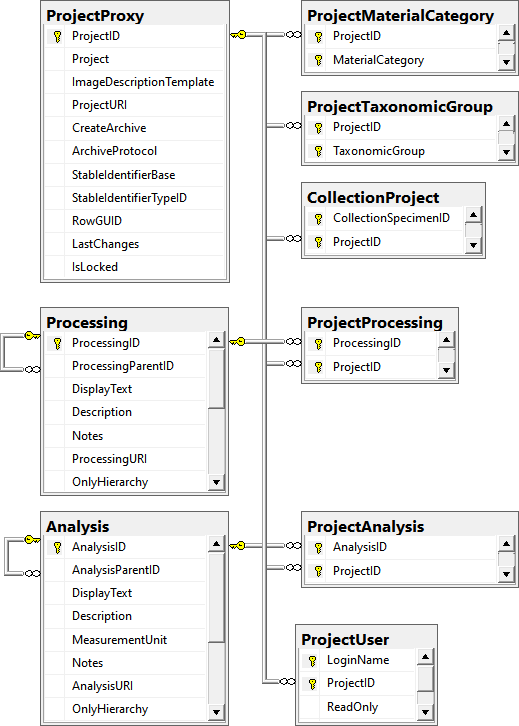
Regulation
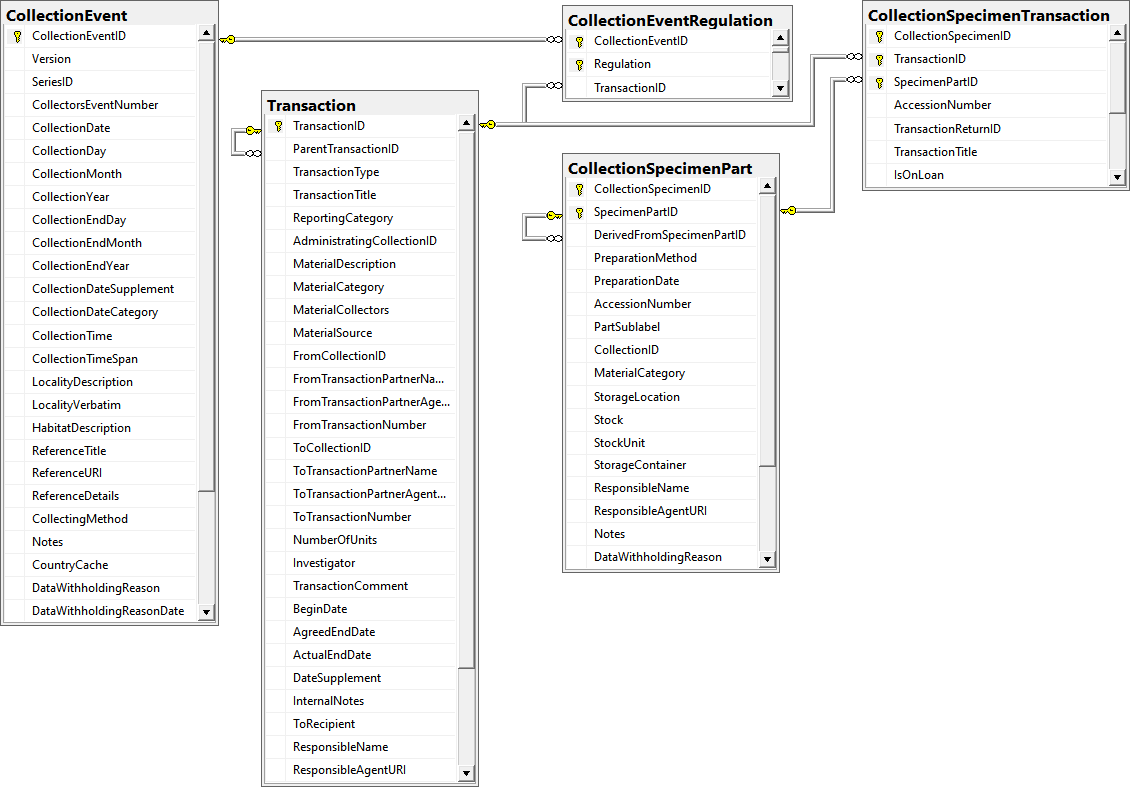
Task
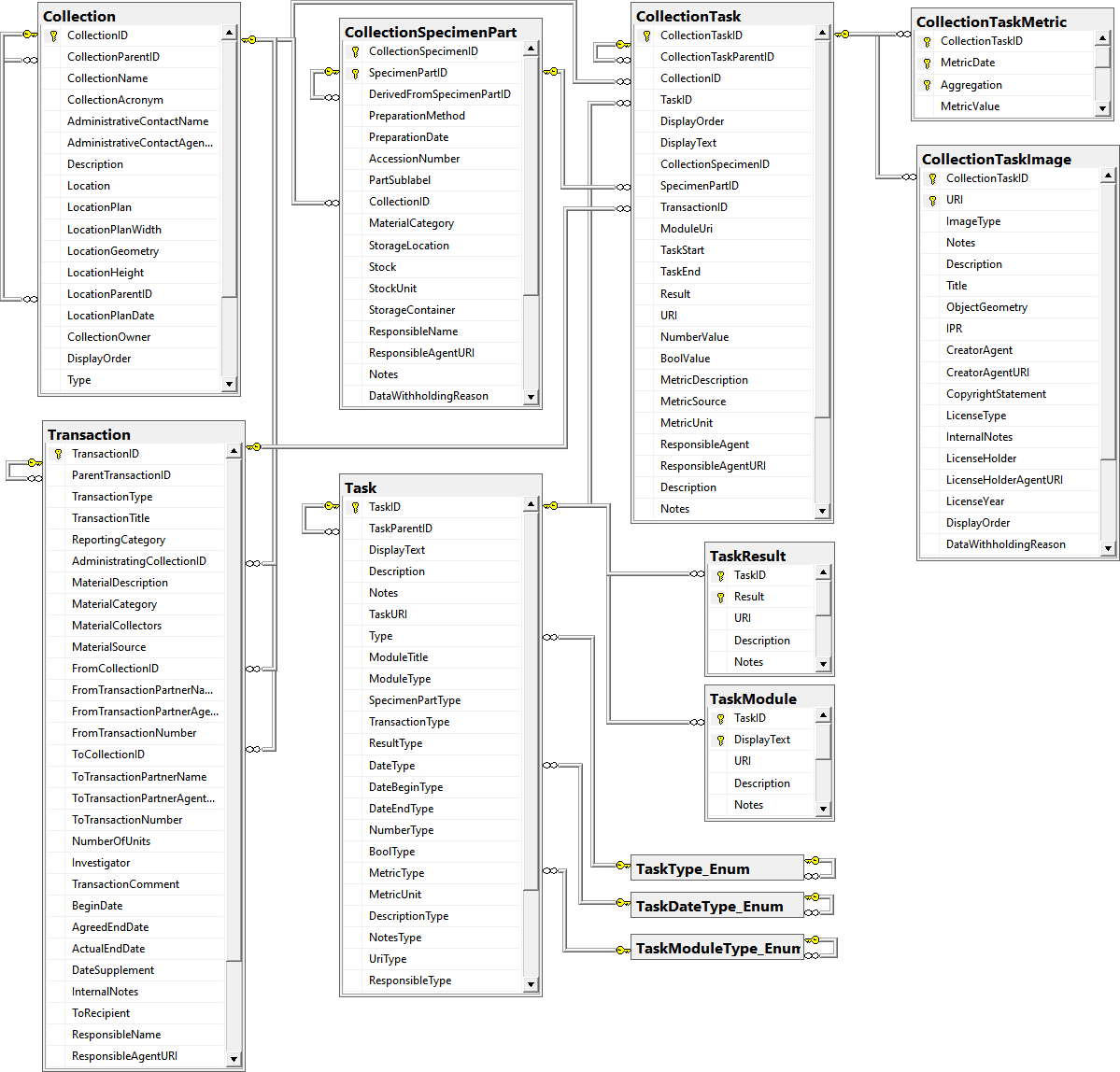
Transaction
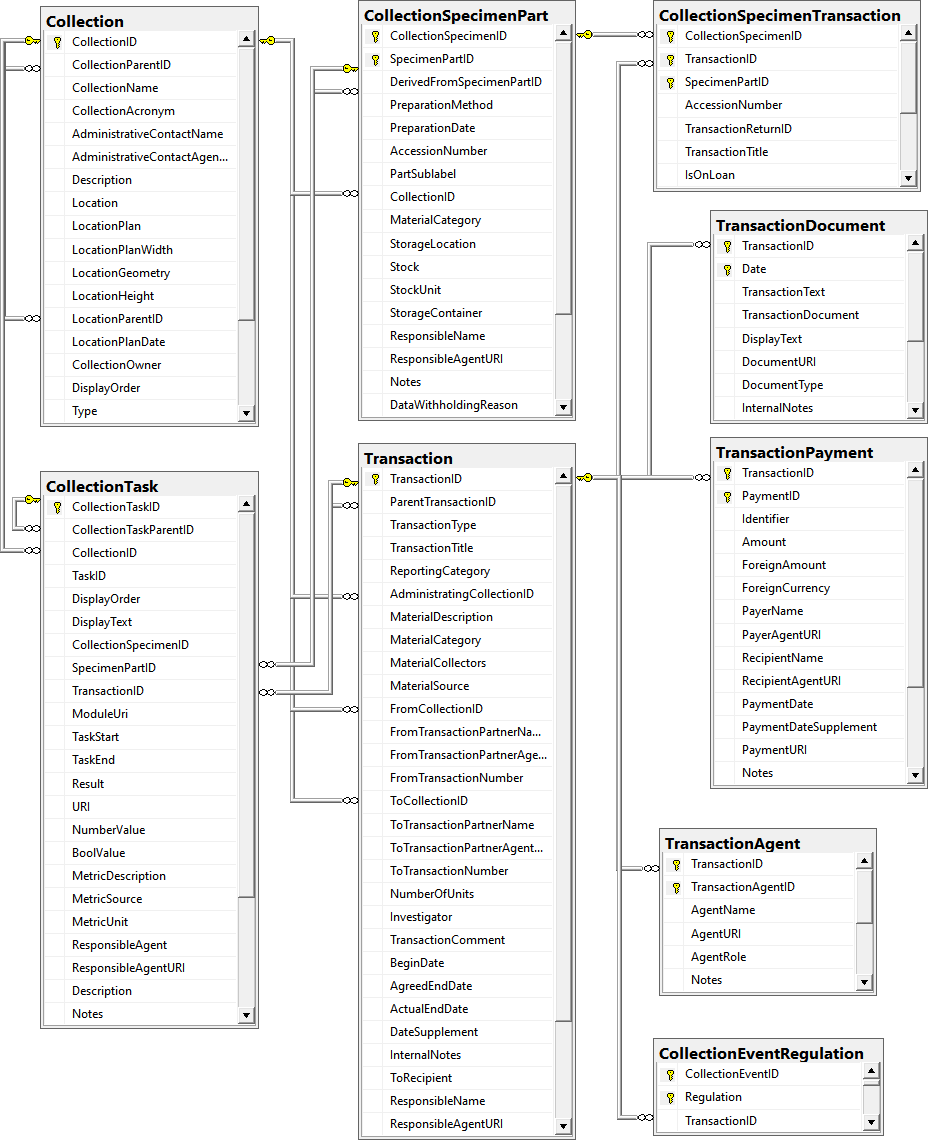
Unit
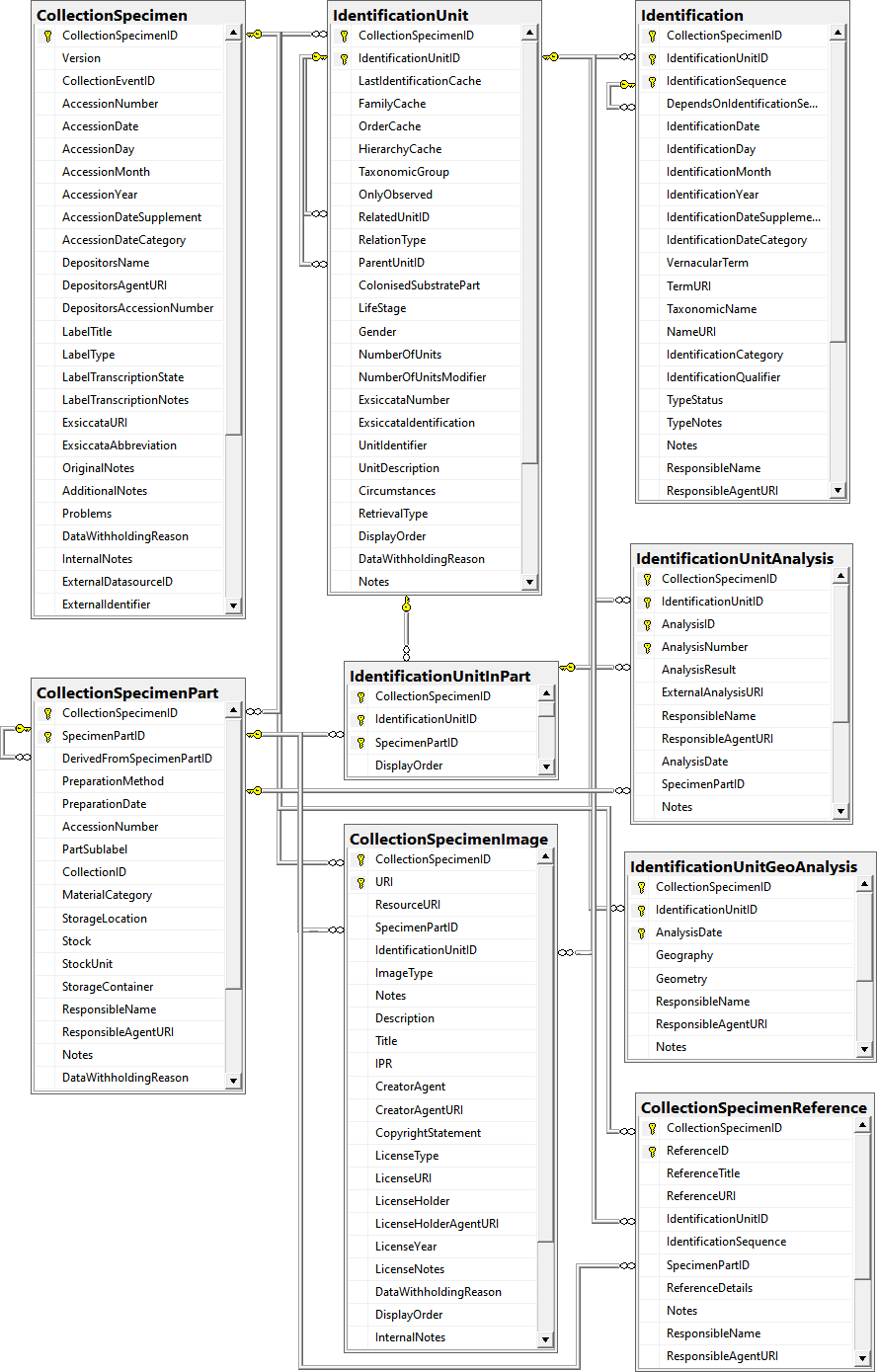
TABLES
Table Analysis
Analysis types used within the database
| Column |
Data type |
Description |
Nullable |
Relation |
| AnalysisID |
int |
ID of the analysis (primary key) |
NO |
- |
| AnalysisParentID |
int |
Analysis ID of the parent analysis, if it belongs to a certain type documented in this table |
YES |
Refers to table Analysis |
| DisplayText |
nvarchar (50) |
Name of the analysis as e.g. shown in user interface |
YES |
- |
| Description |
nvarchar (MAX) |
Description of the analysis |
YES |
- |
| MeasurementUnit |
nvarchar (50) |
The measurement unit used for the analysis, e.g. mm, µmol, kg |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on this analysis |
YES |
- |
| AnalysisURI |
varchar (255) |
URI referring to an external documentation of the analysis |
YES |
- |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries. Default value: (0) |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Table AnalysisResult
Value lists for analysis types with predefined values, e.g. “0, 1, 2, 3, …” for Red list category. Includes description etc. for the values in the list.
| Column |
Data type |
Description |
Nullable |
Relation |
| AnalysisID |
int |
ID of the analysis (primary key) |
NO |
Refers to table Analysis |
| AnalysisResult |
nvarchar (255) |
The categorized value of the analysis |
NO |
- |
| Description |
nvarchar (500) |
Description of enumerated object displayed in the user interface |
YES |
- |
| DisplayText |
nvarchar (50) |
Short abbreviated description of the object displayed in the user interface |
YES |
- |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
YES |
- |
| Notes |
nvarchar (500) |
Internal development notes on usage, definition, etc. of an enumerated object |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user to first enter (typ or import) the data. Default value: suser_sname() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Point in time when the data was first entered (typed or imported) into this database. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data last. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Point in time when this data was updated last. Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table AnalysisTaxonomicGroup
The types of analysis which are available for a taxonomic group
| Column |
Data type |
Description |
Nullable |
Relation |
| AnalysisID |
int |
Analysis ID, foreign key of table Analysis. |
NO |
Refers to table Analysis |
| TaxonomicGroup |
nvarchar (50) |
Taxonomic group the organism, identified by this unit, belongs to. Groups listed in table CollTaxonomicGroup_Enum (= foreign key) |
NO |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table Annotation
Annotations to datasets in the database
| Column |
Data type |
Description |
Nullable |
Relation |
| AnnotationID |
int |
ID of the annotation (primary key) |
NO |
- |
| ReferencedAnnotationID |
int |
If an annotation refers to another annotation, the ID of the referred annotation |
YES |
Refers to table Annotation |
| AnnotationType |
nvarchar (50) |
The type of the annotation as defined in AnnotationType_Enum, e.g. Reference. Default value: N’Annotation' |
NO |
Refers to table AnnotationType_Enum |
| Title |
nvarchar (50) |
Title of the annotation |
YES |
- |
| Annotation |
nvarchar (MAX) |
The annotation entered by the user |
NO |
- |
| URI |
varchar (255) |
The complete URI address of a resource related to the annotation. May be link to a module, e.g. for the annotation type reference |
YES |
- |
| ReferenceDisplayText |
nvarchar (500) |
The title of the reference. If the entry is linked to an external module like DiversityReferences, the cached display text of the referenced data set |
YES |
- |
| ReferenceURI |
varchar (255) |
If the entry is linked to an external module like DiversityReferences, the link to the referenced data set |
YES |
- |
| SourceDisplayText |
nvarchar (500) |
The name of the source. If the entry is linked to an external module like DiversityAgents, the cached display text of the referenced data set |
YES |
- |
| SourceURI |
varchar (255) |
If the entry is linked to an external module like DiversityAgents, the link to the referenced data set |
YES |
- |
| IsInternal |
bit |
If an annotation is restricted to authorized users of the database |
YES |
- |
| ReferencedID |
int |
The ID of the data set in the table the annotation refers to |
NO |
- |
| ReferencedTable |
nvarchar (500) |
The name of the table the annotation refers to |
NO |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
trgInsAnnotation
updating the logging columns
Table AnonymCollector
Anonyms for collectors of whom the names should not be published
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectorsName |
nvarchar (400) |
The name of the collector, PK |
NO |
- |
| Anonymisation |
nvarchar (50) |
The anonymisation phrase for the collector |
YES |
- |
Table CacheDatabase2
Table holding the cache databases connected to the database
| Column |
Data type |
Description |
Nullable |
Relation |
| Server |
varchar (50) |
The name or IP of the server where the cache database is located |
NO |
- |
| DatabaseName |
varchar (50) |
The name of the cache. Default database |
NO |
- |
| Port |
smallint |
The port of the server where the cache database is located |
NO |
- |
| Version |
varchar (50) |
The version of the cache database |
YES |
- |
Table CacheDescription
Table for temperary storage of description of database objects derived e.g. from tables Entity, EntityRepresentation etc.
| Column |
Data type |
Description |
Nullable |
Relation |
| TableName |
varchar (50) |
The name of the table |
NO |
- |
| ColumnName |
varchar (50) |
The name of the table column |
NO |
- |
| LanguageCode |
varchar (50) |
The language code for the description. Default value: ’en-US' |
NO |
- |
| Context |
nvarchar (50) |
A context e.g. as defined in table EntityContext_Enum. Default value: ‘General’ |
NO |
- |
| DisplayText |
nvarchar (50) |
The text for the table or column as shown e.g. in a user interface |
YES |
- |
| Abbreviation |
nvarchar (20) |
The abbreviation for the table or column as shown e.g. in a user interface |
YES |
- |
| Description |
nvarchar (MAX) |
The description for the table column |
YES |
- |
| ID |
int |
A unique ID |
NO |
- |
| Type |
varchar (20) |
Type of the entry. Default value: ‘COLUMN’ |
YES |
- |
| Schema |
varchar (100) |
Schema of the entry. Default value: ‘dbo’ |
YES |
- |
Table Collection
The collections where the specimen are stored
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionID |
int |
Unique reference ID for the collection (= primary key) |
NO |
- |
| CollectionParentID |
int |
For a subcollection within another collection: CollectionID of the collection to which the subcollection belongs. Empty for an independent collection |
YES |
Refers to table Collection |
| CollectionName |
nvarchar (255) |
Name of the collection (e.g. ‘Herbarium Kew’) or subcollection (e.g. ‘cone collection’, ‘alcohol preservations’). This text should be kept relatively short. You may use Description for additional information |
NO |
- |
| CollectionAcronym |
nvarchar (10) |
A unique code for the collection, e.g. the herbarium code from Index Herbariorum |
YES |
- |
| AdministrativeContactName |
nvarchar (500) |
The name of the person or organisation responsible for this collection |
YES |
- |
| AdministrativeContactAgentURI |
varchar (255) |
The URI of the person or organisation responsible for the collection e.g. as provided by the module DiversityAgents |
YES |
- |
| Description |
nvarchar (MAX) |
A short description of the collection |
YES |
- |
| Location |
nvarchar (255) |
Optional location of the collection, e.g. the number within a file system or a description of the room(s) housing the (sub)collection |
YES |
- |
| LocationPlan |
varchar (500) |
URI or file name including path of the floor plan of the collection |
YES |
- |
| LocationPlanWidth |
float |
Width of location plan in meter for calculation of size by provided geometry |
YES |
- |
| LocationGeometry |
geometry (MAX) |
Geometry of the collection within the floor plan |
YES |
- |
| LocationHeight |
float |
Height from ground level, e.g. for the position of sensors |
YES |
- |
| LocationParentID |
int |
If the hierarchy of the location does not match the logical hierarchy, the ID of the parent location |
YES |
Refers to table Collection |
| LocationPlanDate |
datetime |
The date when the plan for the collection has been created |
YES |
- |
| CollectionOwner |
nvarchar (255) |
The owner of the collection as e.g. printed on a label. Should be given if CollectionParentID is null |
YES |
- |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
YES |
- |
| Type |
nvarchar (50) |
Type of the collection, e.g. cupboard, drawer etc. |
YES |
Refers to table CollCollectionType_Enum |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table CollectionAgent
The collector(s) of CollectionSpecimens
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to ID of CollectionEvent (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimen |
| CollectorsName |
nvarchar (255) |
Name of the Collector |
NO |
- |
| CollectorsAgentURI |
varchar (255) |
The URI of the Agent, e.g. as stored within the module DiversityAgents |
YES |
- |
| CollectorsSequence |
datetime2 |
The order of collectors in a team. Automatically set by the database system. Default value: sysdatetime() |
YES |
- |
| CollectorsNumber |
nvarchar (50) |
Number assigned to a specimen or a batch of specimens by the collector during the collection event (= ‘field number’) |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the collector, e.g. if the name is uncertain |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
trgInsCollectionAgent
Setting the version of the dataset
Table CollectionEvent
The event where and when the specimen were collected
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionEventID |
int |
Unique ID for the table CollectionEvent (= primary key) |
NO |
- |
| Version |
int |
The version of the data set. Automatically set by the system. Default value: (1) |
NO |
- |
| SeriesID |
int |
The ID of the related expedition. Relates to the PK of the table CollectionExpedition (foreign key). |
YES |
Refers to table CollectionEventSeries |
| CollectorsEventNumber |
nvarchar (50) |
Number assigned to a collection event by the collector (= ‘field number’) |
YES |
- |
| CollectionDate |
datetime |
The cached date of the collection event calculated from the entries in CollectionDay, -Month and -Year. |
YES |
- |
| CollectionDay |
tinyint |
The day of the date of the event or when the collection event started |
YES |
- |
| CollectionMonth |
tinyint |
The month of the date of the event or when the collection event started |
YES |
- |
| CollectionYear |
smallint |
The year of the date of the event or when the collection event started |
YES |
- |
| CollectionEndDay |
tinyint |
The day of the date of the event or when the collection event ended |
YES |
- |
| CollectionEndMonth |
tinyint |
The month of the date of the event or when the collection event ended |
YES |
- |
| CollectionEndYear |
smallint |
The year of the date of the event or when the collection event ended |
YES |
- |
| CollectionDateSupplement |
nvarchar (100) |
Verbal or additional collection date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. The end date, if the collection event comprises a period. The time of the event, if necessary. |
YES |
- |
| CollectionDateCategory |
nvarchar (50) |
Category of the date of the identification e.g. “system”, “estimated” (= foreign key, see in table CollEventDateCategory_Enum) |
YES |
Refers to table CollEventDateCategory_Enum |
| CollectionTime |
varchar (50) |
The time of the event or when the collection event started |
YES |
- |
| CollectionTimeSpan |
varchar (50) |
The time span e.g. in seconds of the collection event |
YES |
- |
| LocalityDescription |
nvarchar (MAX) |
Locality description of the locality exactly as written on the original label (i.e. without corrections during data entry) |
YES |
- |
| LocalityVerbatim |
nvarchar (MAX) |
Locality as given in historical context, documents and labels |
YES |
- |
| HabitatDescription |
nvarchar (MAX) |
Geo-ecological description of the locality exactly as written on the original label (i.e. without corrections during data entry) |
YES |
- |
| ReferenceTitle |
nvarchar (255) |
The title of the publication where the collection event was published. Note that this is only a cached value where ReferenceURI is present |
YES |
- |
| ReferenceURI |
varchar (255) |
URI (e.g. LSID) of the source publication where the collection event is published, may e.g. refer to the module DiversityReferences |
YES |
- |
| ReferenceDetails |
nvarchar (50) |
The exact location within the reference, e.g. pages, plates |
YES |
- |
| CollectingMethod |
nvarchar (MAX) |
Description of the method used for collecting the samples, e.g. traps, moist chambers, drag net |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the collection event |
YES |
- |
| CountryCache |
nvarchar (50) |
The country where the collection event took place. Cached value derived from an entry in CollectionEventLocalisation |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| DataWithholdingReasonDate |
nvarchar (50) |
The reason for withholding the collection date |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionEventSeries
- CollEventDateCategory_Enum
trgInsCollectionEvent
Setting the date in case of valid date columns
Table CollectionEventImage
The images showing the collection site resp. place of the observations
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionEventID |
int |
Part of primary key, refers to unique ID for the table CollectionEvent (= foreign key) |
NO |
Refers to table CollectionEvent |
| URI |
varchar (255) |
The complete URI address of the image. This is only a cached value, if ResourceID is available and referring to the module DiversityResources |
NO |
- |
| ResourceURI |
varchar (255) |
The URI of the resource (e.g. see module DiversityResources) |
YES |
- |
| ImageType |
nvarchar (50) |
Type of the image, e.g. map |
YES |
Refers to table CollEventImageType_Enum |
| Notes |
nvarchar (MAX) |
Notes to this image concerning the CollectionEvent |
YES |
- |
| Description |
xml (MAX) |
Description of the image |
YES |
- |
| Title |
nvarchar (500) |
Title of the resource |
YES |
- |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
- |
| CreatorAgent |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
- |
| CreatorAgentURI |
varchar (255) |
Link to the module DiversityAgents |
YES |
- |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
- |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e. g. Creative Common Licenses |
YES |
- |
| InternalNotes |
nvarchar (500) |
Internal notes which should not be published e.g. on websites |
YES |
- |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
- |
| LicenseHolderAgentURI |
nvarchar (500) |
The link to a module containing further information on the person or institution holding the license |
YES |
- |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionEvent
- CollEventImageType_Enum
trgInsCollectionEventImage
Setting the version of the dataset
Table CollectionEventLocalisation
The geographic localisation of a CollectionEvent
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionEventID |
int |
Refers to the ID of CollectionEvent (= foreign key and part of primary key) |
NO |
Refers to table CollectionEvent |
| LocalisationSystemID |
int |
Refers to the ID of LocalisationSystem (= foreign key and part of primary key) |
NO |
Refers to table LocalisationSystem |
| Location1 |
nvarchar (255) |
Either a named location selected from a thesaurus (e.g. ‘Germany, Bavaria, Kleindingharting’) or altitude range or other values (e. g. 100-200 m) |
YES |
- |
| Location2 |
nvarchar (255) |
Corresponding value to Location1 e.g. ID or URI of gazetteer or thesaurus |
YES |
- |
| LocationAccuracy |
nvarchar (50) |
The accuracy of the determination of this locality |
YES |
- |
| LocationNotes |
nvarchar (MAX) |
Notes on the location |
YES |
- |
| DeterminationDate |
smalldatetime |
Date of the determination of the geographical localisation |
YES |
- |
| DistanceToLocation |
varchar (50) |
Distance from the specified place to the real location of the collection site (m) |
YES |
- |
| DirectionToLocation |
varchar (50) |
Direction from the specified place to the real location of the collection site (Degrees rel. to north) |
YES |
- |
| ResponsibleName |
nvarchar (255) |
The name of the agent (person or organization) responsible for this entry. |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or organisation responsible for the data (see e.g. module DiversityAgents) |
YES |
- |
| Geography |
geography |
The geography of the localisation |
YES |
- |
| RecordingMethod |
nvarchar (500) |
The method or device used for the recording of the localisation |
YES |
- |
| AverageAltitudeCache |
float |
Calculated altitude as parsed from the location fields |
YES |
- |
| AverageLatitudeCache |
float |
Calculated latitude as parsed from the location fields |
YES |
- |
| AverageLongitudeCache |
float |
Calculated longitude as parsed from the location fields |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionEvent
- LocalisationSystem
trgInsCollectionEventLocalisation
Setting missing geographical values on base of given values
Table CollectionEventMethod
The methods used during a collection event
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionEventID |
int |
Refers to ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionEvent |
| MethodID |
int |
ID of the setting, part of primary key |
NO |
Refers to table Method |
| MethodMarker |
nvarchar (50) |
A marker for the method, part of primary key. Default value: ‘1’ |
NO |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table CollectionEventParameterValue
The values of the parameter of the methods used within a collection event
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionEventID |
int |
Unique ID for the table CollectionEvent (= foreign key and part of primary key) |
NO |
Refers to table CollectionEventMethod |
| MethodID |
int |
ID of the method tool. Referes to table Method (= foreign key and part of primary key) |
NO |
Refers to table CollectionEventMethod and table Parameter |
| MethodMarker |
nvarchar (50) |
A marker for the method, part of primary key. Default value: ‘1’ |
NO |
Refers to table CollectionEventMethod |
| ParameterID |
int |
ID of the parameter tool. Refers to table Parameter (= foreign key and part of primary key) |
NO |
Refers to table Parameter |
| Value |
nvarchar (MAX) |
The value of the parameter, if different of the default value as documented in the table Parameter |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes concerning the value of the parameter |
YES |
- |
| LogInsertedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
YES |
- |
Depending on:
- CollectionEventMethod
- Parameter
Table CollectionEventProperty
A property of a collection site, e.g. exposition, slope, vegetation. May refer to Diversity Workbench module DiversityScientificTerms
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionEventID |
int |
Refers to the ID of CollectionEvent (= foreign key and part of primary key) |
NO |
Refers to table CollectionEvent |
| PropertyID |
int |
The ID of the property of the collection site, foreign key, see table Property |
NO |
Refers to table Property |
| DisplayText |
nvarchar (255) |
The text for the property as shown e.g. in a user interface |
YES |
- |
| PropertyURI |
varchar (255) |
URI referring to an external data source e.g. DiversityTerminology |
YES |
- |
| PropertyHierarchyCache |
nvarchar (MAX) |
A cached text of the complete name of the descriptor including superior categories, if present |
YES |
- |
| PropertyValue |
nvarchar (255) |
The value of a captured feature, e.g. temperature, pH, vegetation etc. If there is a range, this is the lower or first value |
YES |
- |
| ResponsibleName |
nvarchar (255) |
The name of the agent (person or organization) responsible for this entry. |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or organisation responsible for the data (see e.g. module DiversityAgents) |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the property of the collection site. |
YES |
- |
| AverageValueCache |
float |
For numeric values - a cached average value according to the |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
trgInsCollectionEventProperty
Setting the version of the dataset
Table CollectionEventRegulation
Regulation applied to a collection event
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionEventID |
int |
Part of primary key, refers to unique ID for the table CollectionEvent (= foreign key). |
NO |
Refers to table CollectionEvent |
| Regulation |
nvarchar (400) |
Regulation as defined in the table Regulation. Used to ensure, that user checked correct entry with authorized stuff |
NO |
- |
| TransactionID |
int |
Refers to unique TransactionID for the table Transaction (= foreign key) |
YES |
Refers to table Transaction |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionEvent
- Transaction
Table CollectionEventSeries
The series whithin which collection events take place
| Column |
Data type |
Description |
Nullable |
Relation |
| SeriesID |
int |
Primary key. The ID for this series of collection events (= primary key) |
NO |
- |
| SeriesParentID |
int |
The ID of the superior series of collection events |
YES |
Refers to table CollectionEventSeries |
| Description |
nvarchar (MAX) |
The description of the series of collection events as it will be printed on e.g. the label |
NO |
- |
| SeriesCode |
nvarchar (50) |
The user defined code for a series of collection events |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on this series of collection events |
YES |
- |
| Geography |
geography |
The geography of the series of collection events |
YES |
- |
| DateStart |
datetime |
Point in time when the series of collection events started |
YES |
- |
| DateEnd |
datetime |
Point in time when the series of collection events ended |
YES |
- |
| DateCache |
datetime |
The first date of the depending events, used for sorting the expeditions [controlled by the database] |
YES |
- |
| DateSupplement |
nvarchar (100) |
Verbal or additional collection date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. The end date, if the collection event series comprises a period. The time of the event, if necessary. |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Table CollectionEventSeriesDescriptor
The Descriptors for the CollectionEventSeries
| Column |
Data type |
Description |
Nullable |
Relation |
| SeriesID |
int |
Unique ID for the CollectionEventSeries (foreign key + part of primary key) |
NO |
Refers to table CollectionEventSeries |
| DescriptorID |
int |
Unique ID for the descriptor, Part of PK |
NO |
- |
| Descriptor |
nvarchar (200) |
The Descriptor. Default value: '’ |
NO |
- |
| URL |
varchar (500) |
URL of the Descriptor. In case of a module related Descriptor, the link to the module entry resp. the related webservice. Default value: '’ |
YES |
- |
| DescriptorType |
nvarchar (50) |
Type of the Descriptor as described in table CollectionEventSeriesDescriptorType_Enum. Default value: N’Descriptor’ |
YES |
Refers to table CollEventSeriesDescriptorType_Enum |
| LogInsertedBy |
nvarchar (50) |
Name of user who first entered (typed or imported) the data. Default value: suser_sname() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Date and time when the data were first entered (typed or imported) into this database. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of user who last updated the data. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Date and time when the data were last updated. Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionEventSeries
- CollEventSeriesDescriptorType_Enum
Table CollectionEventSeriesImage
The images showing the sites of a series of collection events, e.g. an expedition
| Column |
Data type |
Description |
Nullable |
Relation |
| SeriesID |
int |
Unique ID for the table CollectionEventSeries (= foreign key and part of primary key) |
NO |
Refers to table CollectionEventSeries |
| URI |
varchar (255) |
The complete URI address of the image. This is only a cached value, if ResourceID is available and referring to the module DiversityResources |
NO |
- |
| ResourceURI |
varchar (255) |
The URI of the resource (e.g. see module DiversityResources) |
YES |
- |
| ImageType |
nvarchar (50) |
Type of the image, e.g. map |
YES |
Refers to table CollEventSeriesImageType_Enum |
| Notes |
nvarchar (MAX) |
Notes to this image of the collection site |
YES |
- |
| Description |
xml (MAX) |
Description of the image |
YES |
- |
| Title |
nvarchar (500) |
Title of the resource |
YES |
- |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
- |
| CreatorAgent |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
- |
| CreatorAgentURI |
varchar (255) |
Link to the module DiversityAgents |
YES |
- |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
- |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e.g. Creative Common licenses |
YES |
- |
| InternalNotes |
nvarchar (500) |
Internal notes which should not be published e.g. on websites |
YES |
- |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
- |
| LicenseHolderAgentURI |
nvarchar (500) |
The link to a module containing further information on the person or institution holding the license. |
YES |
- |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionEventSeries
- CollEventSeriesImageType_Enum
Table CollectionExternalDatasource
CollectionExternalDatasource document the sources of the names.
| Column |
Data type |
Description |
Nullable |
Relation |
| ExternalDatasourceID |
int |
An ID to identify an external data collection of the collected specimen (primary key, the ID has no meaning outside of the DiversityWorkbench system) |
NO |
- |
| ExternalDatasourceName |
nvarchar (255) |
The name of the data collection which has been integrated or can be linked to for further analysis |
YES |
- |
| ExternalDatasourceVersion |
nvarchar (255) |
The version of this data collection (either official version number or dates when the collection was integrated) |
YES |
- |
| Rights |
nvarchar (500) |
A description of copyright agreements or permission to use data from the external database |
YES |
- |
| ExternalDatasourceAuthors |
nvarchar (200) |
The persons or institutions responsible for the external database |
YES |
- |
| ExternalDatasourceURI |
nvarchar (300) |
The URI of the database provider or the external database |
YES |
- |
| ExternalDatasourceInstitution |
nvarchar (300) |
The institution responsible for the external database |
YES |
- |
| InternalNotes |
nvarchar (1500) |
Additional notes on this data collection |
YES |
- |
| ExternalAttribute_NameID |
nvarchar (255) |
The table and field name in the external data collection to which CollectionExternalID refers |
YES |
- |
| PreferredSequence |
tinyint |
For selection in e.g. pick lists: of several equal names only the name from the source with the lowest preferred sequence will be provided. |
YES |
- |
| Disabled |
bit |
If this source should be disabled for selection of names e.g. in pick lists. |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Table CollectionImage
The images showing the collection
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionID |
int |
Refers to the ID of Collection (= foreign key and part of primary key) |
NO |
Refers to table Collection |
| URI |
varchar (255) |
The complete URI address of the image. |
NO |
- |
| ImageType |
nvarchar (50) |
Type of the image, e.g. label |
YES |
Refers to table CollCollectionImageType_Enum |
| Notes |
nvarchar (MAX) |
Notes on the collection image |
YES |
- |
| Description |
xml (MAX) |
Description of the image |
YES |
- |
| Title |
nvarchar (500) |
Title of the resource |
YES |
- |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
- |
| CreatorAgent |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
- |
| CreatorAgentURI |
varchar (255) |
Link to the module DiversityAgents |
YES |
- |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
- |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e.g. Creative Common licenses |
YES |
- |
| InternalNotes |
nvarchar (500) |
Internal notes which should not be published e.g. on websites |
YES |
- |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
- |
| LicenseHolderAgentURI |
nvarchar (500) |
The link to a module containing further information on the person or institution holding the license. |
YES |
- |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
- |
| LocationGeometry |
geometry (MAX) |
Geometry of the collection e.g. within a floor plan |
YES |
- |
| RecordingDate |
datetime |
The recording date of the resource |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| LogInsertedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollCollectionImageType_Enum
- Collection
Table CollectionManager
Collection managers within DiversityCollection responsible for specimen transactions
| Column |
Data type |
Description |
Nullable |
Relation |
| LoginName |
nvarchar (50) |
A login name which the user uses to access the DivesityWorkbench, Microsoft domains, etc.. |
NO |
- |
| AdministratingCollectionID |
int |
ID for the collection for which the Manager has the right to administrate the transaction. Corresponds to AdministratingCollectionID in table Transaction. |
NO |
Refers to table Collection |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table CollectionProject
The projects within which the collection specimen were placed
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimen |
| ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
NO |
Refers to table ProjectProxy |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimen
- ProjectProxy
trgInsCollectionProject
Setting LastChanges in table ProjectProxy
Table CollectionSpecimen
The data directly attributed to the collected specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Unique ID for the table CollectionSpecimen (primary key) |
NO |
- |
| Version |
int |
The version of the data set. Default value: (1) |
NO |
- |
| CollectionEventID |
int |
Refers to the ID of table CollectionEvent (= foreign key and part of primary key) |
YES |
Refers to table CollectionEvent |
| AccessionNumber |
nvarchar (50) |
Accession number of the specimen within the collection, e.g. “M-29834752” |
YES |
- |
| AccessionDate |
datetime |
The date of the accession calculated from the entries in AccessionDay, -Month and -Year |
YES |
- |
| AccessionDay |
tinyint |
The day of the date when the specimen was acquired in the collection |
YES |
- |
| AccessionMonth |
tinyint |
The month of the date when the specimen was acquired in the collection |
YES |
- |
| AccessionYear |
smallint |
The year of the date when the specimen was acquired in the collection |
YES |
- |
| AccessionDateSupplement |
nvarchar (255) |
Verbal or additional accession date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
YES |
- |
| AccessionDateCategory |
nvarchar (50) |
Category of the date of the accession e.g. “system”, “estimated” (= foreign key, see in table CollDateCategory_Enum) |
YES |
Refers to table CollDateCategory_Enum |
| DepositorsName |
nvarchar (255) |
The name of the depositor(s) (person or organization responsible for deposition). Where entire collections are deposited, this should also contain the collection name (e.g. ‘Herbarium P. Döbbler’) |
YES |
- |
| DepositorsAgentURI |
varchar (255) |
The URI of the depositor(s) (person or organization responsible for deposition) |
YES |
- |
| DepositorsAccessionNumber |
nvarchar (50) |
Accession number of the specimen within the previous or original collection, e.g. ‘D-23948’ |
YES |
- |
| LabelTitle |
nvarchar (MAX) |
The title of the label e.g. for printing labels. |
YES |
- |
| LabelType |
nvarchar (50) |
Printed, typewritten, typewritten with handwriting added, entirely in handwriting, etc. |
YES |
Refers to table CollLabelType_Enum |
| LabelTranscriptionState |
nvarchar (50) |
The state of the transcription of a label into the database: ‘Not started’, ‘incomplete’, ‘complete’ |
YES |
Refers to table CollLabelTranscriptionState_Enum |
| LabelTranscriptionNotes |
nvarchar (MAX) |
User defined notes on the transcription of the label into the database |
YES |
- |
| ExsiccataURI |
varchar (255) |
If specimen is an exsiccata: The URI of the exsiccata series, e.g. as stored within the DiversityExsiccata module |
YES |
- |
| ExsiccataAbbreviation |
nvarchar (255) |
If specimen is an exsiccata: Standard abbreviation of the exsiccata (not necessarily a unique identifier; editors or publication places may change over time) |
YES |
- |
| OriginalNotes |
nvarchar (MAX) |
Notes found on the label of the specimen by the original collector or from a later revision |
YES |
- |
| AdditionalNotes |
nvarchar (MAX) |
Additional notes made by the editor of the specimen record, e.g. ‘doubtful identification/locality’ |
YES |
- |
| Problems |
nvarchar (255) |
Description of a problem which occurred during data editing. Typically these entries should be deleted after help has been obtained. Do not enter scientific problems here; use AdditionalNotes for such permanent problems! |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null. Default value: N’Withhold by default’ |
YES |
- |
| InternalNotes |
nvarchar (MAX) |
Internal notes which should not be published e.g. on websites |
YES |
- |
| ExternalDatasourceID |
int |
An ID to identify an external data collection of the collected specimen (primary key, the ID has no meaning outside of the DiversityWorkbench system) |
YES |
Refers to table CollectionExternalDatasource |
| ExternalIdentifier |
nvarchar (100) |
The identifier of the external specimen as defined in the external data source |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollDateCategory_Enum
- CollectionEvent
- CollectionExternalDatasource
- CollLabelTranscriptionState_Enum
- CollLabelType_Enum
Table CollectionSpecimenImage
The images of a collection specimen or of an organism (stored in table IdentificationUnit) within this specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimen and table IdentificationUnit |
| URI |
varchar (255) |
The complete URI address of the image. This is only a cached value, if ResourceID is available and referring to the module DiversityResources |
NO |
- |
| ResourceURI |
varchar (255) |
The URI of the image, e.g. as stored in the module DiversityResources. |
YES |
- |
| SpecimenPartID |
int |
Optional: If the data set is not related to a part of a specimen, the ID of a related part (= foreign key) |
YES |
Refers to table CollectionSpecimenPart |
| IdentificationUnitID |
int |
If image refers to only one out of several IdentificationUnits for a specimen, refers to the ID of an IdentificationUnit for a collection specimen (= foreign key) |
YES |
Refers to table IdentificationUnit |
| ImageType |
nvarchar (50) |
Type of the image, e.g. photograph |
YES |
Refers to table CollSpecimenImageType_Enum |
| Notes |
nvarchar (MAX) |
Notes on the specimen image |
YES |
- |
| Description |
xml (MAX) |
Description of the image |
YES |
- |
| Title |
nvarchar (500) |
Title of the resource |
YES |
- |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
- |
| CreatorAgent |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
- |
| CreatorAgentURI |
varchar (255) |
Link to the module DiversityAgents |
YES |
- |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
- |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e. g. Creative Common Licenses |
YES |
- |
| LicenseURI |
varchar (500) |
The URI of the license for the resource |
YES |
- |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
- |
| LicenseHolderAgentURI |
nvarchar (500) |
The link to a module containing further information on the person or institution holding the license. |
YES |
- |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
- |
| LicenseNotes |
nvarchar (500) |
Notice on license for the resource |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| DisplayOrder |
int |
The order in which the images should be shown in an interface. |
YES |
- |
| InternalNotes |
nvarchar (500) |
Internal notes which should not be published e.g. on websites |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimen
- CollectionSpecimenPart
- CollSpecimenImageType_Enum
- IdentificationUnit
CollectionSpecimenImage_URI
Deprecated
trgInsCollectionSpecimenImage
Setting the version of the dataset
Table CollectionSpecimenImageProperty
The properties of images of a collection specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenImage |
| URI |
varchar (255) |
The complete URI address of the image. This is only a cached value, if ResourceID is available and referring to the module DiversityResources |
NO |
Refers to table CollectionSpecimenImage |
| Property |
varchar (255) |
The property of the image |
NO |
- |
| Description |
nvarchar (MAX) |
If description of the property of the image |
YES |
- |
| ImageArea |
geometry (MAX) |
The area in the image the property refers to |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was createdDefault value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
| PropertyURI |
varchar (500) |
The URI of the property of the image, e.g. a link to module DiversityScientificTerms |
YES |
- |
Depending on:
Table CollectionSpecimenPart
Parts of a collected specimen. Includes a possible hierarchy of the parts
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimen and table CollectionSpecimenPart |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen (= part of primary key). |
NO |
- |
| DerivedFromSpecimenPartID |
int |
SpecimenPartID of the specimen from which the current specimen is derived from |
YES |
Refers to table CollectionSpecimenPart |
| PreparationMethod |
nvarchar (MAX) |
The method used for the preparation of the part of the specimen, e.g. the inoculation method for cultures |
YES |
- |
| PreparationDate |
datetime |
Point in time when the part was preparated e.g when it was separated from the source object |
YES |
- |
| AccessionNumber |
nvarchar (50) |
Accession number of the part of the specimen within the collection, if it is different from the accession number of the specimen as stored in the table CollectionSpecimen, e.g. “M-29834752” |
YES |
- |
| PartSublabel |
nvarchar (50) |
The label for a part of a specimen, e.g. “cone”, or a number attached to a duplicate of a specimen |
YES |
- |
| CollectionID |
int |
ID of the collection as stored in table Collection (= foreign key, see table Collection) |
NO |
Refers to table Collection |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc. (= foreign key, see table CollMaterialCategory_Enum). Default value: N’specimen’ |
NO |
Refers to table CollMaterialCategory_Enum |
| StorageLocation |
nvarchar (255) |
A code identifying the place where the specimen is stored within the collection. Frequently the accepted scientific name is used as storage location code. |
YES |
- |
| Stock |
float |
Number of stock units, if the specimen is stored in separated units e.g. several boxes or vessels (max. 255) |
YES |
- |
| StockUnit |
nvarchar (50) |
If empty, the stock is given as a count, else it contains the unit in which stock is expressed, e.g. µl, ml, kg etc. |
YES |
- |
| StorageContainer |
nvarchar (500) |
The container in which the part is stored |
YES |
- |
| ResponsibleName |
nvarchar (255) |
Name of the person or institution responsible for the preparation |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or institution responsible for the preparation (= foreign key) as stored in the module DiversityAgents |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the storage of the sample |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the specimen part is withhold, the reason for withholding the data, otherwise null. |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- Collection
- CollectionSpecimen
- CollMaterialCategory_Enum
trgInsCollectionSpecimenPart
Setting the version of the dataset
Table CollectionSpecimenPartDescription
Description of the specimen part with a standardized vocabulary as defined in the module DiversityScientificTerms
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenPart |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen (= part of primary key). |
NO |
Refers to table CollectionSpecimenPart |
| PartDescriptionID |
int |
ID of the description (Part of primary key) |
NO |
- |
| IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
YES |
- |
| Description |
nvarchar (MAX) |
The description of the part. Cached value if DescriptionTermURI is used. |
YES |
- |
| DescriptionTermURI |
varchar (500) |
Link to an external data source like a web-service or the module DiversityScientificTerms where the description is documented. |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes about this description |
YES |
- |
| DescriptionHierarchyCache |
nvarchar (MAX) |
Hierarchy of the description. For values linked to a module, a cached value provided by the module |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table CollectionSpecimenProcessing
The processing which was applied to a collected specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimen and table CollectionSpecimenPart |
| SpecimenProcessingID |
int |
Unique ID of the processing of a specimen or part of a specimen, part of primary key |
NO |
- |
| ProcessingDate |
datetime |
Point in time of the start of the processing |
YES |
- |
| ProcessingID |
int |
ID of the processing method. Refers to ProcessingID in table Processing (foreign key). Default value: (1) |
NO |
Refers to table Processing |
| Protocoll |
nvarchar (100) |
The label of the processing protocol |
YES |
- |
| SpecimenPartID |
int |
Optional: If the data set is related to a part of a specimen, the ID of a related part (= foreign key, see table CollectionSpecimenPart) |
YES |
Refers to table CollectionSpecimenPart |
| ProcessingDuration |
varchar (50) |
The duration of the processing including the unit (e.g. 5 min) or the end of the processing starting at the processing date (e.g. 23.05.2008) |
YES |
- |
| ResponsibleName |
nvarchar (255) |
Name of the person or institution responsible for the determination |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the processing |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
| ToolUsage |
xml (MAX) |
The tools used for the processing and their usage or settings. |
YES |
- |
Depending on:
- CollectionSpecimen
- CollectionSpecimenPart
- Processing
trgInsCollectionSpecimenProcessing
Setting the version of the dataset
Table CollectionSpecimenProcessingMethod
The methods used for a processing of a specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to ID of CollectionSpecimen (= Foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenProcessing |
| SpecimenProcessingID |
int |
Refers to the ID of the specimen processing (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenProcessing |
| MethodID |
int |
ID of the method, part of primary key |
NO |
Refers to table MethodForProcessing |
| MethodMarker |
nvarchar (50) |
A marker for the method, part of primary key. Default value: ‘1’ |
NO |
- |
| ProcessingID |
int |
ID of the processing. Refers to ProcessingID in table Processing (foreign key). Default value: (1) |
NO |
Refers to table MethodForProcessing |
| LogCreatedWhen |
datetime |
The time when this dataset was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Who created this dataset. Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
datetime |
The last time when this dataset was updated. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Who was the last to update this dataset. Default value: user_name() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimenProcessing
- MethodForProcessing
Table CollectionSpecimenProcessingMethodParameter
The parameter values of a method used for the processing of a specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to ID of CollectionSpecimen (= Foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenProcessingMethod |
| SpecimenProcessingID |
int |
Refers to ID of CollectionSpecimenProcessing (= Foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenProcessingMethod |
| ProcessingID |
int |
ID of the processing. Refers to ProcessingID in table Processing (= Foreign key and part of primary key). Default value: (1) |
NO |
Refers to table CollectionSpecimenProcessingMethod |
| MethodID |
int |
ID of the method (= Foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenProcessingMethod and table Parameter |
| MethodMarker |
nvarchar (50) |
A marker for the method, part of primary key. Default value: ‘1’ |
NO |
Refers to table CollectionSpecimenProcessingMethod |
| ParameterID |
int |
ID of the parameter. Refers to table Parameter (= Foreign key and part of primary key). |
NO |
Refers to table Parameter |
| Value |
nvarchar (MAX) |
The value of the parameter if different of the default value as documented in the table Parameter |
NO |
- |
| LogCreatedWhen |
datetime |
The time when this dataset was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Who created this dataset. Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
datetime |
The last time when this dataset was updated. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Who was the last to update this dataset. Default value: user_name() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimenProcessingMethod
- Parameter
Table CollectionSpecimenReference
A reference related to the collection specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to unique ID of collection specimen record (part of primary key) |
NO |
Refers to table CollectionSpecimen and table IdentificationUnit |
| ReferenceID |
int |
Unique reference ID for the reference record (part of primary key) |
NO |
- |
| ReferenceTitle |
nvarchar (400) |
The title of the publication related to the specimen or parts of it. Note that this is only a cached value where ReferenceURI is present |
NO |
- |
| ReferenceURI |
varchar (500) |
URI of the reference, e.g. a connection to the module DiversityReferences |
YES |
- |
| IdentificationUnitID |
int |
If relation refers to a certain organism within a specimen, the ID of an IdentificationUnit (= foreign key) |
YES |
Refers to table IdentificationUnit |
| IdentificationSequence |
smallint |
Refers to table Identification: The sequence of the identifications. |
YES |
- |
| SpecimenPartID |
int |
If the relation refers to a part of a specimen, the ID of a related part (= foreign key) |
YES |
Refers to table CollectionSpecimenPart |
| ReferenceDetails |
nvarchar (500) |
The exact location within the reference, e.g. pages, plates |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes about the reference |
YES |
- |
| ResponsibleName |
nvarchar (255) |
The name of the agent (person or organization) responsible for this entry. |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or organisation responsible for the data (see e.g. module DiversityAgents) |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimen
- CollectionSpecimenPart
- IdentificationUnit
Table CollectionSpecimenRelation
The relations of a collection specimen to other collection specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Unique reference ID for the collection specimen record (primary key) |
NO |
Refers to table CollectionSpecimen and table IdentificationUnit |
| RelatedSpecimenURI |
varchar (255) |
URI of the related specimen |
NO |
- |
| RelatedSpecimenDisplayText |
varchar (255) |
The name of a related specimen as shown e.g. in a user interface |
NO |
- |
| RelationType |
nvarchar (50) |
Type of the relation between the specimen (= foreign key, see table CollRelationType_Enum) |
YES |
Refers to table CollSpecimenRelationType_Enum |
| RelatedSpecimenCollectionID |
int |
ID of the Collection as stored in table Collection (= foreign key, see table Collection) |
YES |
Refers to table Collection |
| RelatedSpecimenDescription |
nvarchar (MAX) |
Description of the related specimen |
YES |
- |
| IdentificationUnitID |
int |
If relation refers to a certain organism within a specimen, the ID of an IdentificationUnit (= foreign key) |
YES |
Refers to table IdentificationUnit |
| SpecimenPartID |
int |
If the relation refers to a part of a specimen, the ID of a related part (= foreign key) |
YES |
Refers to table CollectionSpecimenPart |
| Notes |
nvarchar (MAX) |
Notes on the relation to the specimen |
YES |
- |
| IsInternalRelationCache |
bit |
If the relation represents a connection between specimen in this database. Default value: (1) |
NO |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- Collection
- CollectionSpecimen
- CollectionSpecimenPart
- CollSpecimenRelationType_Enum
- IdentificationUnit
trgInsCollectionSpecimenRelation
Setting the version of the dataset
Table CollectionSpecimenTransaction
The transactions in which a specimen was involved
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenPart |
| TransactionID |
int |
Unique ID for the table Transaction (= foreign key and part of primary key) |
NO |
Refers to table Transaction |
| SpecimenPartID |
int |
Optional: If the data set is related to a part of a specimen, the ID of a related part (= foreign key, see table CollectionSpecimenPart) |
NO |
Refers to table CollectionSpecimenPart |
| AccessionNumber |
nvarchar (255) |
Accession number which has been assigned to the part of the specimen, e.g. in connection with a former inventory. |
YES |
- |
| TransactionReturnID |
int |
Unique ID for the table Transaction (= foreign key) for the return of a part that has been on loan |
YES |
- |
| TransactionTitle |
nvarchar (200) |
Title as in related table Transaction. Used for validation of correct entry of transaction with type regulation (see insert trigger) |
YES |
- |
| IsOnLoan |
bit |
True, if a specimen is on loan |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of user to first enter (typed or imported) the data. Default value: suser_sname() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Point in time when the data was first entered (typed or imported) into this database. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimenPart
- Transaction
trgInsCollectionSpecimenTransaction
Rollback of regulations if corresponding datasets are missing in table CollectionEventRegulation
Table CollectionTask
A task for a collection. Details are defined in table Task
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionTaskID |
int |
PK of the table |
NO |
- |
| CollectionTaskParentID |
int |
Relation to PK for hierarchy within the data |
YES |
Refers to table CollectionTask |
| CollectionID |
int |
Relation to table Collection. Every ColletionTask needs a relation to a collection |
NO |
Refers to table Collection |
| TaskID |
int |
Relation to table Task where details for the collection task are defined |
NO |
Refers to table Task |
| DisplayOrder |
int |
Display order e.g. in a report. Data with value 0 will not be included. Default value: (1) |
YES |
- |
| DisplayText |
nvarchar (400) |
The display text of the module related data as shown e.g. in a user interface |
YES |
- |
| CollectionSpecimenID |
int |
Refers to the CollectionSpecimenID of CollectionSpecimenPart (= foreign key and part of primary key) |
YES |
Refers to table CollectionSpecimenPart |
| SpecimenPartID |
int |
Unique ID of the part of the collection specimen the task is related to. |
YES |
Refers to table CollectionSpecimenPart |
| TransactionID |
int |
ID of a transaction. Related to PK of table Transaction |
YES |
Refers to table Transaction |
| ModuleUri |
varchar (500) |
The URL of module related data |
YES |
- |
| TaskStart |
datetime |
The start date and or time of the collection tasks. The type is defined in table Task |
YES |
- |
| TaskEnd |
datetime |
The end date and or time of the collection tasks. The type is defined in table Task |
YES |
- |
| Result |
nvarchar (400) |
A text result either taken from a list or entered. The type is defined in table Task |
YES |
- |
| URI |
varchar (500) |
The URI of the collection tasks. The type is defined in table Task |
YES |
- |
| NumberValue |
real |
The numeric value of the collection tasks. The type is defined in table Task |
YES |
- |
| BoolValue |
bit |
The boolean of the collection tasks. The type is defined in table Task |
YES |
- |
| MetricDescription |
nvarchar (500) |
Description of the metric e.g. imported from a time series database like Prometheus |
YES |
- |
| MetricSource |
varchar (4000) |
The source of the metric e.g. the PromQL statement for the import from a timeseries database Prometheus |
YES |
- |
| MetricUnit |
nvarchar (50) |
The unit of the metric, e.g. °C for temperature |
YES |
- |
| ResponsibleAgent |
nvarchar (500) |
The name of the responsible person or institution |
YES |
- |
| ResponsibleAgentURI |
varchar (500) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
YES |
- |
| Description |
nvarchar (MAX) |
The description of the collection tasks. The type is defined in table Task |
YES |
- |
| Notes |
nvarchar (MAX) |
The notes of the collection tasks. The type is defined in table Task |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: dbo.UserID() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: dbo.UserID() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- Collection
- CollectionSpecimenPart
- Task
- Transaction
trgInsCollectionTask
Inserting CollectionID of parent entry if missing
Table CollectionTaskImage
The images showing the CollectionTask
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionTaskID |
int |
Refers to the ID of CollectionTask (= foreign key and part of primary key) |
NO |
Refers to table CollectionTask |
| URI |
varchar (255) |
The complete URI address of the image. |
NO |
- |
| ImageType |
nvarchar (50) |
Type of the image, e.g. label |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the CollectionTask image |
YES |
- |
| Description |
xml (MAX) |
Description of the image |
YES |
- |
| Title |
nvarchar (500) |
Title of the resource |
YES |
- |
| ObjectGeometry |
geometry (MAX) |
The geometry of an object placed within the image |
YES |
- |
| IPR |
nvarchar (500) |
Intellectual Property Rights; the rights given to persons for their intellectual property |
YES |
- |
| CreatorAgent |
nvarchar (500) |
Person or organization originally creating the resource |
YES |
- |
| CreatorAgentURI |
varchar (255) |
Link to the module DiversityAgents |
YES |
- |
| CopyrightStatement |
nvarchar (500) |
Notice on rights held in and for the resource |
YES |
- |
| LicenseType |
nvarchar (500) |
Type of an official or legal permission to do or own a specified thing, e.g. Creative Common licenses |
YES |
- |
| InternalNotes |
nvarchar (500) |
Internal notes which should not be published e.g. on websites |
YES |
- |
| LicenseHolder |
nvarchar (500) |
The person or institution holding the license |
YES |
- |
| LicenseHolderAgentURI |
nvarchar (500) |
The link to a module containing further information on the person or institution holding the license. |
YES |
- |
| LicenseYear |
nvarchar (50) |
The year of license declaration |
YES |
- |
| DisplayOrder |
smallint |
The display order of the image |
YES |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| LogInsertedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: dbo.UserID() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: dbo.UserID() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table CollectionTaskMetric
The metric related to a collection task
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionTaskID |
int |
Refers to the ID of CollectionTask (= foreign key and part of primary key) |
NO |
Refers to table CollectionTask |
| MetricDate |
datetime |
Date and time of the metric, part of PK |
NO |
- |
| Aggregation |
nvarchar (50) |
The Aggregation applied for retrieval of the value, e.g. max, avg etc.. Default value: N’none’ |
NO |
Refers to table CollTaskMetricAggregation_Enum |
| MetricValue |
real |
The value of the metric |
YES |
- |
| LogInsertedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: dbo.UserID() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: dbo.UserID() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionTask
- CollTaskMetricAggregation_Enum
Table CollectionUser
Users of collections within DiversityCollection
| Column |
Data type |
Description |
Nullable |
Relation |
| LoginName |
nvarchar (50) |
A login name which the user uses to access the DivesityWorkbench, Microsoft domains, etc.. |
NO |
- |
| CollectionID |
int |
ID for the collection for the user has access to administrate the transaction. |
NO |
Refers to table Collection |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table Entity
The entities in an application e.g. the tables and columns in a database
| Column |
Data type |
Description |
Nullable |
Relation |
| Entity |
varchar (500) |
The name of the entity, e.g. Table.Column.Content within the database or a unique string for e.g. a message within the DiversityWorkbench e.g. “DiversityWorkbench.Message.Connection.NoAccess”, PK |
NO |
- |
| DisplayGroup |
nvarchar (50) |
If DiversityWorkbench entities should be displayed in a group, the name of the group |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the entity |
YES |
- |
| Obsolete |
bit |
True if an entity is obsolete. Obsolete entities may be kept to ensure compatibility with older modules |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Table EntityRepresentation
The description of the entity in a certain context in different languages
| Column |
Data type |
Description |
Nullable |
Relation |
| Entity |
varchar (500) |
The name of the entity. Foreign key, relates to table Entity |
NO |
Refers to table Entity |
| LanguageCode |
nvarchar (50) |
ISO 639: 2-letter codes for the language of the content |
NO |
Refers to table EntityLanguageCode_Enum |
| EntityContext |
nvarchar (50) |
The context for the representation, e.g. “Exchange with ABCD”, “collection management” or “observation” as defined in table EntityContext_Enum |
NO |
Refers to table EntityContext_Enum |
| DisplayText |
nvarchar (50) |
The text for the entity as shown e.g. in a user interface |
YES |
- |
| Abbreviation |
nvarchar (20) |
The abbreviation for the entity as shown e.g. in a user interface |
YES |
- |
| Description |
nvarchar (MAX) |
The description of the entity |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the representation of the entity |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- Entity
- EntityContext_Enum
- EntityLanguageCode_Enum
Table EntityUsage
The usage of an entity in a certain context, e.g. hidden, readonly
| Column |
Data type |
Description |
Nullable |
Relation |
| Entity |
varchar (500) |
The name of the entity. Foreign key, relates to table Entity |
NO |
Refers to table Entity |
| EntityContext |
nvarchar (50) |
The context for the representation, e.g. “Exchange with ABCD”, “collection management” or “observation” as defined in table EntityContext_Enum |
NO |
Refers to table EntityContext_Enum |
| Accessibility |
nvarchar (50) |
If the access of entity is restricted to e.g. read only or it can be edited without restrictions. |
YES |
Refers to table EntityAccessibility_Enum |
| Determination |
nvarchar (50) |
If a value is determined e.g. by the system or the user |
YES |
Refers to table EntityDetermination_Enum |
| Visibility |
nvarchar (50) |
If the entity is visible or hidden from e.g. a user interface |
YES |
Refers to table EntityVisibility_Enum |
| PresetValue |
nvarchar (500) |
If a value is preset, the value or SQL statement for the value, e.g. ‘determination’ for identifications when using a mobile device during an expedition |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the usage of the entity |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- Entity
- EntityAccessibility_Enum
- EntityContext_Enum
- EntityDetermination_Enum
- EntityVisibility_Enum
Table ExternalIdentifier
An external identier related to a dataset, e.g. a DOI
| Column |
Data type |
Description |
Nullable |
Relation |
| ID |
int |
ID of the identifier (Primary key) |
NO |
- |
| ReferencedTable |
nvarchar (128) |
The name of the table the external identifier refers to |
NO |
- |
| ReferencedID |
int |
The ID of the data set in the table the external identifier refers to |
NO |
- |
| Type |
nvarchar (50) |
The type of the identifier as defined in table ExternalIdentifierType |
YES |
Refers to table ExternalIdentifierType |
| Identifier |
nvarchar (500) |
The identifier |
YES |
- |
| URL |
varchar (500) |
A URL with further informations about the identifier |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes about the identifier |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table ExternalIdentifierType
The type of an external identier, e.g. DOI
| Column |
Data type |
Description |
Nullable |
Relation |
| Type |
nvarchar (50) |
The type of external identifiers (primary key) |
NO |
- |
| ParentType |
nvarchar (50) |
The superior type of this type |
YES |
- |
| URL |
varchar (500) |
A URL providing further informations about this type |
YES |
- |
| Description |
nvarchar (MAX) |
The description of this type |
YES |
- |
| InternalNotes |
nvarchar (MAX) |
Internal notes about the type |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newid() |
NO |
- |
Table Identification
The identifications of the organisms within a specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table Identification and table IdentificationUnit |
| IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
NO |
Refers to table Identification and table IdentificationUnit |
| IdentificationSequence |
smallint |
The sequence of the identifications. The last identification (having the highest sequence) is regarded as valid. Default value: (1) |
NO |
- |
| DependsOnIdentificationSequence |
smallint |
if the identification depends on another identification, e.g. for rocks where several terms from a terminology should be included |
YES |
Refers to table Identification |
| IdentificationDate |
datetime |
The date of the identification calculated from the entries in IdentificationDay, -Month and -Year |
YES |
- |
| IdentificationDay |
tinyint |
The day of the identification |
YES |
- |
| IdentificationMonth |
tinyint |
The month of the identification |
YES |
- |
| IdentificationYear |
smallint |
The year of the identification. The year may be empty if only the day or month are known. |
YES |
- |
| IdentificationDateSupplement |
nvarchar (255) |
Verbal or additional identification date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’ |
YES |
- |
| IdentificationDateCategory |
nvarchar (50) |
Category of the date of the identification e.g. “system”, “estimated” (= foreign key, see in table CollDateCategory_Enum) |
YES |
Refers to table CollIdentificationDateCategory_Enum |
| VernacularTerm |
nvarchar (255) |
Name or term other than a taxonomic (= scientific) name, e.g. ‘pine’, ’limestone’, ‘conifer’, ‘hardwood’ |
YES |
- |
| TermURI |
varchar (500) |
The URI of the term, e.g. as provided by the module DiversityScientificTerms. |
YES |
- |
| TaxonomicName |
nvarchar (255) |
Valid name of the species (including the taxonomic author where available). Example: ‘Rosa canina L.’ |
YES |
- |
| NameURI |
varchar (255) |
The URI of the taxonomic name, e.g. as provided by the module DiversityTaxonNames. |
YES |
- |
| IdentificationCategory |
nvarchar (50) |
Category of the identification e.g. ‘determination’, ‘confirmation’, ‘absence’ (= foreign key, see table CollIdentificationCategory_Enum) |
YES |
Refers to table CollIdentificationCategory_Enum |
| IdentificationQualifier |
nvarchar (50) |
Qualification of the identification e.g. “cf.”," aff.", “sp. nov.” (= foreign key, see table CollIdentificationQualifier_Enum) |
YES |
Refers to table CollIdentificationQualifier_Enum |
| TypeStatus |
nvarchar (50) |
If identification unit is type of a taxonomic name: holotype, syntype, etc. (= foreign key, see table CollTypeStatus_Enum) |
YES |
Refers to table CollTypeStatus_Enum |
| TypeNotes |
nvarchar (MAX) |
Notes on the typification of this specimen |
YES |
- |
| Notes |
nvarchar (MAX) |
User defined notes, e.g. the reason for a re-determination / change of the name, etc. |
YES |
- |
| ResponsibleName |
nvarchar (255) |
Name of the person or institution responsible for the determination |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollIdentificationCategory_Enum
- CollIdentificationDateCategory_Enum
- CollIdentificationQualifier_Enum
- CollTypeStatus_Enum
- IdentificationUnit
trgIdentificationInsert
Updating the LastIdentificationCache in IdentificationUnit
trgInsIdentification
Updating empty date columns depending on a given date
Table IdentificationUnit
Organism which is present in or on a collectied specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimen and table IdentificationUnit |
| IdentificationUnitID |
int |
ID of the IdentificationUnit (= part of primary key). Usually one of possibly several organisms present on the collected specimen. Example: parasite with hyperparasite on plant leaf = 3 units, |
NO |
- |
| LastIdentificationCache |
nvarchar (255) |
The last identification as entered in table Identification |
NO |
- |
| FamilyCache |
nvarchar (255) |
A cached value of the family of the taxon of the last identification. Can be set by the editor, if NameURI in table Identification is NULL, otherwise set by the system. |
YES |
- |
| OrderCache |
nvarchar (255) |
A cached value of the order of the taxon of the last identification. Can be set by the editor, if NameURI in table Identification is NULL, otherwise set by the system. |
YES |
- |
| HierarchyCache |
nvarchar (500) |
A cached value fo the superior taxonomy of the last identification as derived from a taxonomic data provider |
YES |
- |
| TaxonomicGroup |
nvarchar (50) |
Taxonomic group the organism, identified by this unit, belongs to. Groups listed in table CollTaxonomicGroup_Enum (= foreign key) |
NO |
Refers to table CollTaxonomicGroup_Enum |
| OnlyObserved |
bit |
True, if the organism was only observed rather than collected. It is therefore not present on the preserved specimen. Example: Tree under which the collected mycorrhizal fungus grew. Default value: (0) |
YES |
- |
| RelatedUnitID |
int |
The IdentificationUnitID of the organism or substrate on which this organism is growing (= foreign key) |
YES |
Refers to table IdentificationUnit |
| RelationType |
nvarchar (50) |
The relation of a unit to its substrate, e.g. parasitism, symbiosis etc. as stored in CollRelationType_Enum (= foreign key) |
YES |
Refers to table CollUnitRelationType_Enum |
| ParentUnitID |
int |
The IdentificationUnitID of a parent organism of which this organism is a child of (= foreign key). |
YES |
Refers to table IdentificationUnit |
| ColonisedSubstratePart |
nvarchar (255) |
If a substrate association exists: part of the substrate which is affected in the interaction (e.g. ’leaves’, if a fungus is growing on the leaves of an infected plant) |
YES |
- |
| LifeStage |
nvarchar (255) |
Examples: ‘II, III’ for spore generations of rusts or ‘seed’, ‘seedling’ etc. for higher plants |
YES |
- |
| Gender |
nvarchar (50) |
The sex of the organism, e.g. ‘female’ |
YES |
- |
| NumberOfUnits |
smallint |
The number of units of this organism, e.g. 400 beetles in a bottle |
YES |
- |
| NumberOfUnitsModifier |
nvarchar (100) |
A modifier for the number of units of this organism, e.g. ca. 400 beetles in a bottle |
YES |
- |
| ExsiccataNumber |
nvarchar (50) |
If specimen is an exsiccata: Number of current specimen within the exsiccata series |
YES |
- |
| ExsiccataIdentification |
smallint |
Refers to the IdentificationSequence in Identification (= foreign key). The name under which the collectied specimen or this organism is published within an exsiccata. |
YES |
- |
| UnitIdentifier |
nvarchar (50) |
An identifier for the identification of the unit, e.g. a number painted on a tree within an experimental plot |
YES |
- |
| UnitDescription |
nvarchar (50) |
Description of the unit, especially if not an organism but parts or remnants of it were present or observed, e.g. a nest of an insect or a song of a bird |
YES |
- |
| Circumstances |
nvarchar (50) |
Circumstances of the occurence of the organism |
YES |
Refers to table CollCircumstances_Enum |
| RetrievalType |
nvarchar (50) |
The way the data about the unit were retrieved, e.g. observation, literature |
YES |
Refers to table CollRetrievalType_Enum |
| DisplayOrder |
smallint |
The sequence in which the units within this specimen will appear on e.g. a label where the first unit may be printed in the header and others in the text below. 0 means the unit should not appear on a label. Default value: (1) |
NO |
- |
| DataWithholdingReason |
nvarchar (255) |
If the data set is withhold, the reason for withholding the data, otherwise null |
YES |
- |
| Notes |
nvarchar (MAX) |
Further information on the organism or interaction, e.g. infection symptoms like ‘producing galls’ |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollCircumstances_Enum
- CollectionSpecimen
- CollRetrievalType_Enum
- CollTaxonomicGroup_Enum
- CollUnitRelationType_Enum
trgInsIdentificationUnit
setting the display oder for the new unit to the next number if none or an already present on was given
Table IdentificationUnitAnalysis
The analysis values taken from an organism resp. object
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table IdentificationUnit and table IdentificationUnitInPart |
| IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
NO |
Refers to table IdentificationUnit and table IdentificationUnitInPart |
| AnalysisID |
int |
Analysis ID, foreign key of table Analysis. |
NO |
Refers to table Analysis |
| AnalysisNumber |
nvarchar (50) |
Number of the analysis |
NO |
- |
| AnalysisResult |
nvarchar (MAX) |
The result of the analysis |
YES |
- |
| ExternalAnalysisURI |
varchar (255) |
An URI for an analysis as defined in an external datasoure |
YES |
- |
| ResponsibleName |
nvarchar (255) |
Name of the person or institution responsible for the determination |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
YES |
- |
| AnalysisDate |
nvarchar (50) |
The date of the analysis |
YES |
- |
| SpecimenPartID |
int |
ID of the part of a specimen (optional, foreign key) if the analysis was done with a part of the specimen (see table CollectionSpecimenPart). |
YES |
Refers to table IdentificationUnitInPart |
| Notes |
nvarchar (MAX) |
Notes on this analysis |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
| ToolUsage |
xml (MAX) |
The tools used for the analysis and their usage or settings. |
YES |
- |
Depending on:
- Analysis
- IdentificationUnit
- IdentificationUnitInPart
trgInsIdentificationUnitAnalysis
Setting the version of the dataset
Table IdentificationUnitAnalysisMethod
The methods used for an analysis
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to ID of CollectionSpecimen (= Foreign key and part of primary key) |
NO |
Refers to table IdentificationUnitAnalysis |
| IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
NO |
Refers to table IdentificationUnitAnalysis |
| MethodID |
int |
ID of the method, part of primary key |
NO |
Refers to table MethodForAnalysis |
| MethodMarker |
nvarchar (50) |
A marker for the method, part of primary key. Default value: ‘1’ |
NO |
- |
| AnalysisID |
int |
ID of the processing. Refers to AnalysisID in table Processing (foreign key). Default value: (1) |
NO |
Refers to table IdentificationUnitAnalysis and table MethodForAnalysis |
| AnalysisNumber |
nvarchar (50) |
Number of the analysis |
NO |
Refers to table IdentificationUnitAnalysis |
| LogCreatedWhen |
datetime |
The time when this dataset was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Who created this dataset. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
The last time when this dataset was updated. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Who was the last to update this dataset. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- IdentificationUnitAnalysis
- MethodForAnalysis
Table IdentificationUnitAnalysisMethodParameter
The parameter values of a method used for an analysis
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to ID of CollectionSpecimen (= Foreign key and part of primary key) |
NO |
Refers to table IdentificationUnitAnalysisMethod |
| IdentificationUnitID |
int |
ID of the identification unit (= Foreign key and part of primary key) |
NO |
Refers to table IdentificationUnitAnalysisMethod |
| AnalysisID |
int |
ID of the analysis. Refers to AnalysisID in table Analysis (= Foreign key and part of primary key). Default value: (1) |
NO |
Refers to table IdentificationUnitAnalysisMethod |
| AnalysisNumber |
nvarchar (50) |
Number of the analysis (= Foreign key and part of primary key) |
NO |
Refers to table IdentificationUnitAnalysisMethod |
| MethodID |
int |
ID of the method (= Foreign key and part of primary key) |
NO |
Refers to table IdentificationUnitAnalysisMethod and table Parameter |
| MethodMarker |
nvarchar (50) |
A marker for the method, part of primary key. Default value: ‘1’ |
NO |
Refers to table IdentificationUnitAnalysisMethod |
| ParameterID |
int |
ID of the parameter tool. Refers to table Parameter (= Foreign key and part of primary key). |
NO |
Refers to table Parameter |
| Value |
nvarchar (MAX) |
The value of the parameter if different of the default value as documented in the table Parameter |
YES |
- |
| LogCreatedWhen |
datetime |
The time when this dataset was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Who created this dataset. Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
datetime |
The last time when this dataset was updated. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Who was the last to update this dataset. Default value: user_name() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- IdentificationUnitAnalysisMethod
- Parameter
Table IdentificationUnitGeoAnalysis
The geographical position or region of an organism at a certain time
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table IdentificationUnit |
| IdentificationUnitID |
int |
Refers to the ID of IdentficationUnit (= foreign key and part of primary key) |
NO |
Refers to table IdentificationUnit |
| AnalysisDate |
datetime |
The date of the analysis |
NO |
- |
| Geography |
geography |
The geography where the organism resp. object was located according to WGS84, e.g. a point (latitude, longitude and altitude). |
YES |
- |
| Geometry |
geometry (MAX) |
The geometry of the place the organism resp. object was observed, e.g. an area |
YES |
- |
| ResponsibleName |
nvarchar (255) |
Name of the person or institution responsible for the determination |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
URI of the person or institution responsible for the determination (= foreign key) as stored in the module DiversityAgents. |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on this analysis |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table IdentificationUnitInPart
The list of the organisms which are found in a part of the specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| CollectionSpecimenID |
int |
Refers to the ID of CollectionSpecimen (= foreign key and part of primary key) |
NO |
Refers to table CollectionSpecimenPart and table IdentificationUnit |
| IdentificationUnitID |
int |
ID of the identification unit in table IdentificationUnit (= part of primary key). Usually one of possibly several organisms present on the collection specimen. Example: parasite with hyperparasite on plant leaf = 3 units, |
NO |
Refers to table IdentificationUnit |
| SpecimenPartID |
int |
ID of the part of a specimen (optional, foreign key), if the identification unit is located on a part of the specimen (see table CollectionSpecimenPart). |
NO |
Refers to table CollectionSpecimenPart |
| DisplayOrder |
smallint |
The sequence in which the units within this part will appear on e.g. a label where the first unit may be printed in the header and others in the text below. 0 means the unit should not appear on a label. Default value: (1) |
NO |
- |
| LogInsertedBy |
nvarchar (50) |
Name of the user to first enter (typ or import) the data. Default value: suser_sname() |
YES |
- |
| LogInsertedWhen |
smalldatetime |
Point in time when the data was first entered (typed or imported) into this database. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data last. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
smalldatetime |
Point in time when this data was updated last. Default value: getdate() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollectionSpecimenPart
- IdentificationUnit
trgInsIdentificationUnitInPart
Setting the version of the dataset
Table LocalisationSystem
The geographic localisation systems, e.g. coordinates
| Column |
Data type |
Description |
Nullable |
Relation |
| LocalisationSystemID |
int |
Unique ID for the localisation system (= Primary key) |
NO |
- |
| LocalisationSystemParentID |
int |
LocalisationSystemID of the superior LocalisationSystem |
YES |
Refers to table LocalisationSystem |
| LocalisationSystemName |
nvarchar (100) |
Name of the system used for the determination of the place of the collection, e. g. Gauss-Krüger, MTB, GIS |
NO |
- |
| DefaultAccuracyOfLocalisation |
nvarchar (50) |
The default for the accuracy of values which can be reached with this method |
YES |
- |
| DefaultMeasurementUnit |
nvarchar (50) |
The default measurement unit for the localisation system, e.g. m, geographic coordinates. |
YES |
- |
| ParsingMethodName |
nvarchar (50) |
Internal value, specifying a programming method used for parsing text in fields Location1/Location2 in table CollectionLocalisation |
YES |
- |
| DisplayText |
nvarchar (50) |
Short abbreviated description of the localisation system as displayed in the user interface |
YES |
- |
| DisplayEnable |
bit |
Specifies, if this item is enabled to be used within the database. Localisation systems can be disabled to avoid seeing them, but keep the definition for the future. |
YES |
- |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
YES |
- |
| Description |
nvarchar (255) |
Description of the localisation method |
YES |
- |
| DisplayTextLocation1 |
nvarchar (50) |
Short abbreviated description of the attribute Location1 in the table CollectionGeography as displayed in the user interface |
YES |
- |
| DescriptionLocation1 |
nvarchar (255) |
Description of the attribute Location1 in the table CollectionGeography as displayed in the user interface |
YES |
- |
| DisplayTextLocation2 |
nvarchar (50) |
Short abbreviated description of the attribute Location2 in the table CollectionGeography as displayed in the user interface |
YES |
- |
| DescriptionLocation2 |
nvarchar (255) |
Description of the attribute Location2 in the table CollectionGeography as displayed in the user interface |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Table Method
Methods used within the database
| Column |
Data type |
Description |
Nullable |
Relation |
| MethodID |
int |
ID of the Method (Primary key) |
NO |
- |
| MethodParentID |
int |
MethodID of the parent Method, if it belongs to a certain type documented in this table |
YES |
Refers to table Method |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries. Default value: (0) |
YES |
- |
| DisplayText |
nvarchar (50) |
Name of the Method as e.g. shown in user interface |
YES |
- |
| Description |
nvarchar (MAX) |
Description of the Method |
YES |
- |
| MethodURI |
varchar (255) |
URI referring to an external documentation of the Method |
YES |
- |
| ForCollectionEvent |
bit |
If a method may be used during a collection event |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on this method |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Table MethodForAnalysis
Methods available for a Analysis
| Column |
Data type |
Description |
Nullable |
Relation |
| AnalysisID |
int |
ID of the table Analysis (foreign key and part of primary key) |
NO |
Refers to table Analysis |
| MethodID |
int |
ID of the table Method (foreign key and part of primary key) |
NO |
Refers to table Method |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table MethodForProcessing
Methods available for a processing
| Column |
Data type |
Description |
Nullable |
Relation |
| ProcessingID |
int |
ID of the table Processing (foreign key and part of primary key) |
NO |
Refers to table Processing |
| MethodID |
int |
ID of the table Method (foreign key and part of primary key) |
NO |
Refers to table Method |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table Parameter
The variable parameters within a method
| Column |
Data type |
Description |
Nullable |
Relation |
| MethodID |
int |
ID of the Method (foreign key and part of primary key) |
NO |
Refers to table Method |
| ParameterID |
int |
ID of the Parameter (part of primary key) |
NO |
- |
| DisplayText |
nvarchar (50) |
Name of the parameter as e.g. shown in user interface |
YES |
- |
| Description |
nvarchar (MAX) |
Description of the parameter |
YES |
- |
| ParameterURI |
varchar (255) |
URI referring to an external documentation of the Parameter |
YES |
- |
| DefaultValue |
nvarchar (MAX) |
The default value of the parameter |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on this parameter |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table Processing
The processings of the specimen
| Column |
Data type |
Description |
Nullable |
Relation |
| ProcessingID |
int |
ID of the processing (primary key) |
NO |
- |
| ProcessingParentID |
int |
The ID of the superior type of the processing |
YES |
Refers to table Processing |
| DisplayText |
nvarchar (50) |
The display text of the processing as shown e.g. in a user interface |
YES |
- |
| Description |
nvarchar (MAX) |
Description of the processing |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the processing |
YES |
- |
| ProcessingURI |
varchar (255) |
A URI for a processing as defined in an external data source |
YES |
- |
| OnlyHierarchy |
bit |
If the entry is only used for the hierarchical arrangement of the entries. Default value: (0) |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Table ProcessingMaterialCategory
The processings which are possible for a certain material category
| Column |
Data type |
Description |
Nullable |
Relation |
| ProcessingID |
int |
ID of the processing. Refers to ProcessingID in table Processing (foreign key). Default value: (1) |
NO |
Refers to table Processing |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc.. Default value: N’specimen’ |
NO |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table ProjectAnalysis
The types of the analysis which are available for a project
| Column |
Data type |
Description |
Nullable |
Relation |
| AnalysisID |
int |
ID of the analysis (primary key) |
NO |
Refers to table Analysis |
| ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
NO |
Refers to table ProjectProxy |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table ProjectMaterialCategory
The material categorys which are possible for a certain Project
| Column |
Data type |
Description |
Nullable |
Relation |
| ProjectID |
int |
ID of the Project. Refers to ProjectID in table ProjectProxy (foreign key). Default value: (1) |
NO |
Refers to table ProjectProxy |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc.. Default value: N’specimen’ |
NO |
Refers to table CollMaterialCategory_Enum |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollMaterialCategory_Enum
- ProjectProxy
Table ProjectProcessing
The types of processing available within a project
| Column |
Data type |
Description |
Nullable |
Relation |
| ProcessingID |
int |
ID of the table Processing (foreign key and part of primary key) |
NO |
Refers to table Processing |
| ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
NO |
Refers to table ProjectProxy |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table ProjectTaxonomicGroup
The taxonomic group which are possible for a certain Project
| Column |
Data type |
Description |
Nullable |
Relation |
| ProjectID |
int |
ID of the Project. Refers to ProjectID in table ProjectProxy (foreign key). Default value: (1) |
NO |
Refers to table ProjectProxy |
| TaxonomicGroup |
nvarchar (50) |
Taxonomic group of specimen. Examples: ‘plant’, ‘animal’, etc.. Default value: N’plant’ |
NO |
Refers to table CollTaxonomicGroup_Enum |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- CollTaxonomicGroup_Enum
- ProjectProxy
Table ProjectUser
The projects which a user can access
| Column |
Data type |
Description |
Nullable |
Relation |
| LoginName |
nvarchar (50) |
A login name which the user uses to access the DivesityWorkbench, Microsoft domains, etc.. |
NO |
Refers to table UserProxy |
| ProjectID |
int |
ID of the project to which the specimen belongs (Projects are defined in DiversityProjects) |
NO |
Refers to table ProjectProxy |
| ReadOnly |
bit |
If the user has only read access to data of this project. Default value: (0) |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
trgInsProjectUser
Setting ReadOnly in dependence of locked projects
Table Property
The list of the properties that can be specified for the collection site
| Column |
Data type |
Description |
Nullable |
Relation |
| PropertyID |
int |
Unique ID for the property (= primary key) |
NO |
- |
| PropertyParentID |
int |
PropertyID of the superior property |
YES |
Refers to table Property |
| PropertyName |
nvarchar (100) |
Name of the system used for the description of the collection site, e.g. Chronostratigraphy |
NO |
- |
| DefaultAccuracyOfProperty |
nvarchar (50) |
The default for the accuracy of values which can be reached with this method |
YES |
- |
| DefaultMeasurementUnit |
nvarchar (50) |
The default measurement unit for the characterisation system, e.g. pH |
YES |
- |
| ParsingMethodName |
nvarchar (50) |
Internal value, specifying a programming method used for parsing text in table CollectionEventProperty |
NO |
- |
| DisplayText |
nvarchar (50) |
Short abbreviated description of the site property as displayed in the user interface |
YES |
- |
| DisplayEnabled |
bit |
Specifies, if this item is enabled to be used within the database. Properties can be disabled to avoid seeing them, but keep the definition for the future. Default value: (1) |
YES |
- |
| DisplayOrder |
smallint |
The order in which the entries are displayed. The order may be changed at any time, but all values must be unique. |
YES |
- |
| Description |
nvarchar (MAX) |
Description of the property |
YES |
- |
| PropertyURI |
varchar (1000) |
The URI of the property, e.g. as provided by the module DiversityScientificTerms. |
YES |
- |
| PropertyType |
nvarchar (50) |
Type of the collection site property, e.g. Chronostratigraphy |
YES |
Refers to table PropertyType_Enum |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table ReplicationPublisher
Databases providing data via replication
| Column |
Data type |
Description |
Nullable |
Relation |
| DatabaseName |
varchar (255) |
The name of the publishing database |
NO |
- |
| Server |
varchar (255) |
The name or address of the server where the publishing database is located |
NO |
- |
| Port |
smallint |
The port used by the server |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
Table Task
The Tasks of the collection
| Column |
Data type |
Description |
Nullable |
Relation |
| TaskID |
int |
ID of the Task (primary key) |
NO |
- |
| TaskParentID |
int |
The ID of the superior type of the Task |
YES |
Refers to table Task |
| DisplayText |
nvarchar (50) |
The display text of the Task as shown e.g. in a user interface |
YES |
- |
| Description |
nvarchar (MAX) |
Description of the Task |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes on the Task |
YES |
- |
| TaskURI |
varchar (500) |
A URI for a Task as defined in an external data source |
YES |
- |
| Type |
nvarchar (50) |
The type of the task as defined in table TaskType_Enum. Default value: ‘Task’ |
YES |
Refers to table TaskType_Enum |
| ModuleTitle |
nvarchar (50) |
The title for module related data for collection tasks. Not available in depending collection task if empty |
YES |
- |
| ModuleType |
nvarchar (50) |
The DiversityWorkbench module to which a task is related. Not available in depending collection task if empty |
YES |
Refers to table TaskModuleType_Enum |
| SpecimenPartType |
nvarchar (50) |
The description of the collection specimen part to which a task is related. Not available in depending collection task if empty |
YES |
- |
| TransactionType |
nvarchar (50) |
The description of the transaction to which a task is related. Not available in depending collection task if empty |
YES |
- |
| ResultType |
nvarchar (50) |
The display text for the results as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| DateType |
nvarchar (50) |
The date and time details defined for a task. Not available in depending collection task if empty |
YES |
Refers to table TaskDateType_Enum |
| DateBeginType |
nvarchar (50) |
The definition of the begin for date and time details defined for a task. Not available in depending collection task if empty |
YES |
- |
| DateEndType |
nvarchar (50) |
The definition of the end for date and time details defined for a task. Not available in depending collection task if empty |
YES |
- |
| NumberType |
nvarchar (50) |
The definition for the numeric value as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| BoolType |
nvarchar (50) |
The definition for the boolean value as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| MetricType |
nvarchar (50) |
The definition for the metric as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| MetricUnit |
nvarchar (50) |
The unit of the metric, e.g. °C for temperature |
YES |
- |
| DescriptionType |
nvarchar (50) |
The definition for the description as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| NotesType |
nvarchar (50) |
The definition for the notes as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| UriType |
nvarchar (50) |
The definition for the URI as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| ResponsibleType |
nvarchar (50) |
The definition for the responsible agent as shown in a user interface. Not available in depending collection task if empty |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: dbo.UserID() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: dbo.UserID() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- TaskDateType_Enum
- TaskModuleType_Enum
- TaskType_Enum
Table TaskModule
Data from DiversityWorkbench modules used used within a task
| Column |
Data type |
Description |
Nullable |
Relation |
| TaskID |
int |
ID of the task, part of PK, relates to PK of table Task |
NO |
Refers to table Task |
| DisplayText |
nvarchar (400) |
Display text as provided by the module, part of PK |
NO |
- |
| URI |
varchar (500) |
URI linking the dataset of the module |
YES |
- |
| Description |
nvarchar (MAX) |
Optional description of the linked data, e.g. the common name for taxa |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes related to the dataset |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
ID of the creator of this data set. Default value: dbo.UserID() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
ID of the person to update this data set last. Default value: dbo.UserID() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table TaskResult
List or results provided for the CollectionTask of the corresponding type
| Column |
Data type |
Description |
Nullable |
Relation |
| TaskID |
int |
ID of the task, part of PK, relates to PK of table Task |
NO |
Refers to table Task |
| Result |
nvarchar (400) |
The result, part of PK |
NO |
- |
| URI |
varchar (500) |
A URI of the result providing further information |
YES |
- |
| Description |
nvarchar (MAX) |
The description of the entry |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes about the entry |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
ID of the creator of this data set. Default value: dbo.UserID() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
ID of the person to update this data set last. Default value: dbo.UserID() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table Transaction
Transactions like loan, borrow, gift, exchange etc. of specimen, if they are e.g. permanently or temporary transfered from one collection to another
| Column |
Data type |
Description |
Nullable |
Relation |
| TransactionID |
int |
Unique ID for the transaction (= primary key) |
NO |
- |
| ParentTransactionID |
int |
The ID of a preceding transaction of a superior transaction, if transactions are organized in a hierarchy. |
YES |
Refers to table Transaction |
| TransactionType |
nvarchar (50) |
Type of the transaction, e.g. gift in or out, exchange in or out, purchase in or out. Default value: N’exchange’ |
NO |
Refers to table CollTransactionType_Enum |
| TransactionTitle |
nvarchar (200) |
The title of the transaction as e.g. shown in an user interface |
NO |
- |
| ReportingCategory |
nvarchar (50) |
A group defined for the transaction, e.g. a taxonomic group as used for exchange balancing |
YES |
- |
| AdministratingCollectionID |
int |
ID of the collection which is responsible for the administration of the transaction. |
NO |
Refers to table Collection |
| MaterialDescription |
nvarchar (MAX) |
Description of the material of this transaction. Default value: '’ |
YES |
- |
| MaterialCategory |
nvarchar (50) |
Material category of specimen. Examples: ‘herbarium sheets’, ‘drawings’, ‘microscopic slides’ etc.. Default value: N’specimen’ |
YES |
- |
| MaterialCollectors |
nvarchar (MAX) |
The collectors of the material |
YES |
- |
| MaterialSource |
nvarchar (500) |
The source of the material within a transaction, e.g. an excavation. |
YES |
- |
| FromCollectionID |
int |
The ID of the collection from which the specimen were transferred, e.g. the donating collection of a gift. |
YES |
Refers to table Collection |
| FromTransactionPartnerName |
nvarchar (255) |
Name of the person or institution from which the specimen were transferred, e.g. the donator of a gift. |
YES |
- |
| FromTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
YES |
- |
| FromTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the source of the specimen, e.g. the donating collection of a gift |
YES |
- |
| ToCollectionID |
int |
The ID of the collection to which the specimen were transferred, e.g. the receiver of a gift. |
YES |
Refers to table Collection |
| ToTransactionPartnerName |
nvarchar (255) |
Name of the person or institution to which the specimen were transferred, e.g. the receiver of a gift |
YES |
- |
| ToTransactionPartnerAgentURI |
varchar (255) |
The URI of the transaction partner (see e.g. module DiversityAgents) |
YES |
- |
| ToTransactionNumber |
nvarchar (50) |
Number or code by which a transaction may be recorded by the administration of the destination of the specimen, e.g. the receiving collection of a gift |
YES |
- |
| NumberOfUnits |
int |
The number of units which were (initially) included in the transaction |
YES |
- |
| Investigator |
nvarchar (200) |
The investigator for whose study a transacted material was sent |
YES |
- |
| TransactionComment |
nvarchar (MAX) |
Comments on the exchanged material addressed to the transaction partner |
YES |
- |
| BeginDate |
datetime |
Date when the transaction started |
YES |
- |
| AgreedEndDate |
datetime |
End of the transaction period, e.g. if the time for borrowing the specimen is restricted |
YES |
- |
| ActualEndDate |
datetime |
Actual end of the transaction after a prolongation when e.g. the date of return for a loan was prolonged by the owner. |
YES |
- |
| DateSupplement |
nvarchar (100) |
Verbal or additional date information, e.g. ’end of summer 1985’, ‘first quarter’, ‘1888-1892’. |
YES |
- |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction not to be published e.g. on a web page |
YES |
- |
| ToRecipient |
nvarchar (255) |
The recipient receiving the transaction e.g. if not derived from the link to DiversityAgents |
YES |
- |
| ResponsibleName |
nvarchar (255) |
The person responsible for this transaction |
YES |
- |
| ResponsibleAgentURI |
varchar (255) |
The URI of the person, team or organisation responsible for the data (see e.g. module DiversityAgents) |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: user_name() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: user_name() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
- Collection
- CollTransactionType_Enum
Table TransactionAgent
Agents involved in the transaction
| Column |
Data type |
Description |
Nullable |
Relation |
| TransactionID |
int |
Unique ID for the transaction, refers to table Transaction (= part of primary key and foreign key) |
NO |
Refers to table Transaction |
| TransactionAgentID |
int |
Unique ID for the Agent within the transaction (= part of primary key) |
NO |
- |
| AgentName |
nvarchar (500) |
Name of the person or institution |
YES |
- |
| AgentURI |
varchar (500) |
Link to the source for further informations about the agent, e.g in the module DiversityAgents |
YES |
- |
| AgentRole |
nvarchar (500) |
Role of the agent within the transaction |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes about the agent |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
The standard text phrases for transactions
| Column |
Data type |
Description |
Nullable |
Relation |
| Comment |
nvarchar (400) |
Text as transferred into the comment of a transaction |
NO |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Table TransactionDocument
The history of transactions or the documents connected to the transactions
| Column |
Data type |
Description |
Nullable |
Relation |
| TransactionID |
int |
Unique ID for the transaction, refers to table Transaction (= part of primary key and foreign key) |
NO |
Refers to table Transaction |
| Date |
datetime |
The date of the event of a transaction |
NO |
- |
| TransactionText |
nvarchar (MAX) |
The text of a transaction document |
YES |
- |
| TransactionDocument |
image |
A scanned document connected to this transaction |
YES |
- |
| DisplayText |
nvarchar (255) |
A display text as shown e.g. in a user interface to characterize the document |
YES |
- |
| DocumentURI |
varchar (1000) |
A link to a web resource of the document the document |
YES |
- |
| DocumentType |
nvarchar (255) |
The type of the document |
YES |
- |
| InternalNotes |
nvarchar (MAX) |
Internal notes on this transaction |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Table TransactionPayment
The payments within a transaction
| Column |
Data type |
Description |
Nullable |
Relation |
| TransactionID |
int |
Unique ID for the transaction, refers to table Transaction (= part of primary key and foreign key) |
NO |
Refers to table Transaction |
| PaymentID |
int |
Unique ID for the payment (= part of primary key) |
NO |
- |
| Identifier |
nvarchar (500) |
An identifier for the payment like a booking number or invoice number. |
YES |
- |
| Amount |
float |
Amount of the payment in the default currency as defined in TransactionCurrency |
YES |
- |
| ForeignAmount |
float |
If the payment was not in the default currency as defined in TransactionCurrency, the amount of the payment in foreign currency. |
YES |
- |
| ForeignCurrency |
nvarchar (50) |
If the payment was not in the default currency as defined in TransactionCurrency, the foreign currency of the payment |
YES |
- |
| PayerName |
nvarchar (500) |
Name of the person or institution paying the amount |
YES |
- |
| PayerAgentURI |
varchar (500) |
Link to the source for further information about the payer, e.g. in the module DiversityAgents. |
YES |
- |
| RecipientName |
nvarchar (500) |
Agent receiving the payment |
YES |
- |
| RecipientAgentURI |
varchar (500) |
Link to the source for further information about the recipient of the payment, e.g. in the module DiversityAgents. |
YES |
- |
| PaymentDate |
datetime |
Date of the payment |
YES |
- |
| PaymentDateSupplement |
nvarchar (50) |
Supplement to the date of the payment, e.g. if the original date is not a real date like ‘summer 1920’ or ‘1910 - 1912’ |
YES |
- |
| PaymentURI |
varchar (500) |
A link to an external administration system for the payment |
YES |
- |
| Notes |
nvarchar (MAX) |
Notes about the payment |
YES |
- |
| LogCreatedWhen |
datetime |
Point in time when this data set was created. Default value: getdate() |
YES |
- |
| LogCreatedBy |
nvarchar (50) |
Name of the creator of this data set. Default value: suser_sname() |
YES |
- |
| LogUpdatedWhen |
datetime |
Point in time when this data set was updated last. Default value: getdate() |
YES |
- |
| LogUpdatedBy |
nvarchar (50) |
Name of the person to update this data set last. Default value: suser_sname() |
YES |
- |
| RowGUID |
uniqueidentifier |
-. Default value: newsequentialid() |
NO |
- |
Depending on:
Login administration
To administrate the logins on the database server, their permissions and
roles respectively as well as access to projects choose
Administration - Database - Logins ... from the menu. A window
will open as shown below.
Statistics
To see the activity of a login click on the  button. A window will open as shown below listing all databases and
tables together with the timespan (From - To) and the number of data
sets where any activity of the current login has been found.
button. A window will open as shown below listing all databases and
tables together with the timespan (From - To) and the number of data
sets where any activity of the current login has been found.
To see the current activity on the server click on the
 button. A window as shown below will open
listing all user related processes on the server.
button. A window as shown below will open
listing all user related processes on the server.
Creation of login
To create a new login click on the  button. A
window will open as shown below.
button. A
window will open as shown below.
Here you can enter the name of the new login, the password and the
information about the user which will be stored in a DiversityAgents
database. You may either create a new entry in this database or select
an existing one: Click on the  DWB button
to search for a name in the database (see below).
DWB button
to search for a name in the database (see below).
Copy a login
To copy a login including all permissions etc. into a new login, select
the original login in the list and click on the
 button.
button.
Edit a login
To edit the access for a login on the server select the login in the
list. If a login should be disabled  , uncheck
the enabled checkbox (see below).
, uncheck
the enabled checkbox (see below).

All databases on the server will be listed with the current database
showing a yellow background. The databases where the login has no
access will be listed in gray while the databases accessible for a
login are black.

Access of a login to a database
To allow the access to a database select the database from the list and
choose database as shown below.

Roles of a login in a database
Use the > and < buttons to add or remove roles for the login
in the database (see below).
Projects for a login in a database
Depending on the database you can edit the list of projects accessible
for a login (see below).
There are 4 states of accessibility for projects
 Full access: The user can edit the
data
Full access: The user can edit the
data Read only access: The user can only
read the data
Read only access: The user can only
read the data Locked: The project is locked. Nobody
can change the data
Locked: The project is locked. Nobody
can change the data No access: The user has no access via a
project
No access: The user has no access via a
project
Projects are related to the module DiversityProjects. To get additional
information about a project select it in the the list and click on the
 button.
button.
To load additional projects click on the Load projects
 button. A window will open as shown below.
Projects already in the database will be listed in
green, missing projects in red (see below). Check all projects you need in your database and
click the Start download
button. A window will open as shown below.
Projects already in the database will be listed in
green, missing projects in red (see below). Check all projects you need in your database and
click the Start download  button.
button.
Overview for a login
If you see an overview of all permissions and project for a login, click
on the  button. A window a shown below will
open. It lists all
button. A window a shown below will
open. It lists all  modules and their
modules and their
 databases, the
databases, the  roles,
roles,
 accessible projects and
accessible projects and
 read only projects for a login.
read only projects for a login.
To copy the permissions and projects of the current login to another
login, select the login where the settings should be copied to from the
list at the base of the window and click on the  button to copy the settings for all databases or the
button to copy the settings for all databases or the
 button to copy the settings of the selected
database into this login.
button to copy the settings of the selected
database into this login.
Overview for a database
If you see an overview of all user and roles in a database, click on the
 button. A window a shown below will open.
It lists all
button. A window a shown below will open.
It lists all  user and
user and  roles
in the database.
roles
in the database.
To remove a user, select it in the list and click on the
 button.
button.
Correction of logins
If you select one of the databases, at the base a
 button may appear. This indicates, that
there are windows logins listed where the name of the login does not
match the logins of the server. This may happen if e.g. a database was
moved from one server to another. To correct this, click on the button.
A list of deviating logins will be shown, that can be corrected
automatically.
button may appear. This indicates, that
there are windows logins listed where the name of the login does not
match the logins of the server. This may happen if e.g. a database was
moved from one server to another. To correct this, click on the button.
A list of deviating logins will be shown, that can be corrected
automatically.
If logins with the same name but different server are found, one of them
has to be deleted to make the correction possible. You will get a list
where you can select those that should be removed.
Select the duplicate logins that should be removed and click OK.
These are the tools to handle the basic parts of the database. These
tools are only available for the owner of the database and should be
handled with care as any changes in the database may disable
the connection of your client to the database. Before changing any parts
of the database it is recommended to backup the current state
of the database. To use these tools, choose Administation → Database
→  Database tools ... from the menu. A
window will open as shown below.
Database tools ... from the menu. A
window will open as shown below.
Description
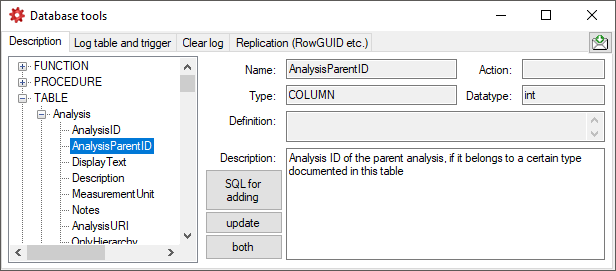
The Description section (see above) shows the basic definitions of
the objects in the database and enables you to enter a description for
these objects including tables and their columns, functions and their
parameter etc. With the buttons SQL for adding, update and
both you can generate SQL statements for the creation of the
descriptions in your database. Use the button both if you are not
sure if a description is already present as it will generate a SQL
statement working with existing and missing descriptions (see below).
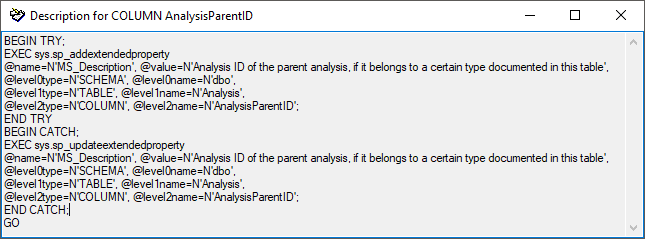
The button  Fill Cache fills the
table CacheDescription where all descriptions are collected for easy
access.
Fill Cache fills the
table CacheDescription where all descriptions are collected for easy
access.
Log table and trigger
In the Log table and trigger section (see below) click on the List
tables button to see all tables within the database. The Table
section shows the basic definitions of a selected table. If columns for
logging the date and responsible user for inserting and updating the
data are missing, you can use the Attach ... button to attach these
columns to the table. Furthermore you may add a RowGUID to the table
as e.g. a preparation for a replication.
In the Log table section (see below) you can create a logging table
for the selected table in a format as used within the Diversity
Workbench. Click on the Show SQL ... button to show the
SQL-statement that will create the logging table. If an old logging
table should be kept, choose the Keep old log table option. If your
table should support the version setting from a main table, choose the
Add the column LogVersion option. To finally create the logging
table click on the Create LogTable ... button.
The triggers for insert, update and delete are created
in the according sections (see below). If an old trigger exists, its
definition will be shown in the upper part of the window. Click on the
Show SQL button to see the definition of the trigger according to
the current definition of the table in a format as used in the Diversity
Workbench. If a trigger should set the version in a main table, which
the current table is related to, choose the Add version setting to
trigger option. To enable this option you must select the version
table first. To finally create the trigger click on the Create
trigger button. The update and delete triggers will transfer the
original version of the data into the logging tables as defined above,
where you can inspect the history of the data sets.
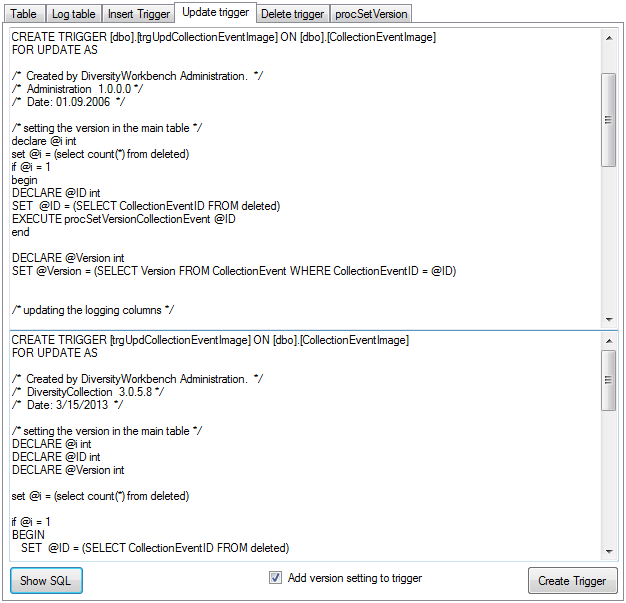
If so far no procedure for setting the version in a main table is
defined, you can create this procedure in the last section. Click on the
Show SQL button to see the definition and on the Create Procedure button
to create the procedure (see below).
Preparation for replication
If you want to use replication within you module, the tables need
certain columns and a log table. These preparations can be performed by
a script, generated in the section Replication (see below). Select the
tables you want to include in the process and create the script. This
script can than be included in an update of the database. Please ensure
that these changes are only be done by expert staff.
Clear logtables
If for any reason you want to clear the log tables of the database, this
can be done in the Clear log tab as shown below. Click on the List
tables button to list the log tables. Then select those that should be
cleared and click on the Clear log of selected tables
 button (see below). Please keep in mind that
any restoration of data from the log is only possible as long as the
data can be retrieved from the log.
button (see below). Please keep in mind that
any restoration of data from the log is only possible as long as the
data can be retrieved from the log.
Data protection
To implement the  General Data Protection Regulation of the European
Union several steps have to be performed in a database:
General Data Protection Regulation of the European
Union several steps have to be performed in a database:
- Generate a skript using this tool (see below) to convert all tables
and insert objects according to the requirements:
- Add columns ID, PrivacyConsent and PrivacyConsentDate in table
UserProxy
- Grant update to PrivacyConsent and PrivacyConsentDate in table
UserProxy
- Create update trigger for UserProxy setting the
PrivacyConsentDate
- Create the function providing the ID of the user from UserProxy
- Create the function providing the name of the user from
UserProxy
- Create the function PrivacyConsentInfo providing common
information
- For every table:
- Insert users from a table into UserProxy (if missing so far)
- Insert users from the corresponding log table into UserProxy
(if missing so far)
- Change the constraints for the logging columns (User_Name()
→ UserID())
- Replace user name with ID in logging columns
- Replace user name with ID in logging columns of the log
table
- Adapt description of the logging columns
- Include the skript in an update of the database
- Check the database for update triggers, functions using e.g.
CURRENT_USER, USER_NAME, SUSER_SNAME etc. where user names must be
replaced with their IDs. Create a script performing these tasks and
include it into an update for the database
- Adapt the client to the now changed informations (e.g. query for
responsible etc.)
After these changes the only place where the name of a user is stored is
the table UserProxy together with the ID. Removing the name (see below)
will remove any information about the user leaving only a number linked
to the information within depending data.
To generate a script for the objects and changes needed to implement the
General Data Protection
Regulation
use the  Data protection tab as shown below. The
generated script will handle the standard objects (logging columns) but
not any additional circumstances within the database. For these you need
to inspect the database in detail and create a script to handle them on
your own.
Data protection tab as shown below. The
generated script will handle the standard objects (logging columns) but
not any additional circumstances within the database. For these you need
to inspect the database in detail and create a script to handle them on
your own.
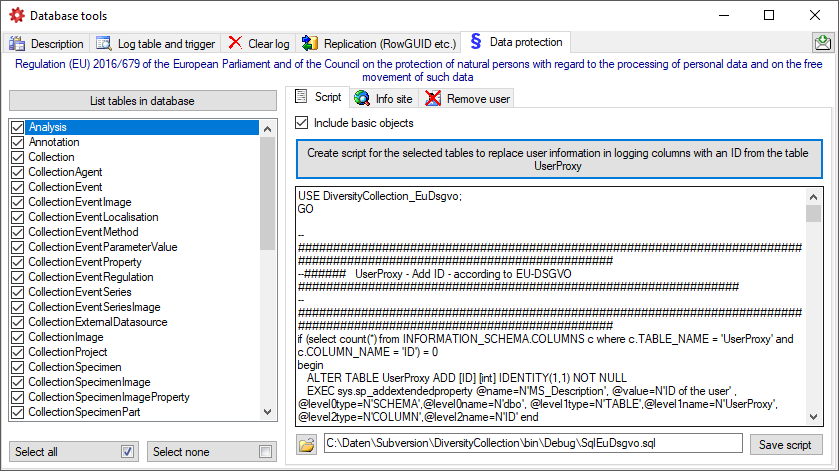
To set the website where detailed information about the handling of the
General Data Protection
Regulation
within the DiversityWorkbench resp. the current database is provided,
click on the  button on the
button on the  Info site tab. If unchanged, the default
site
for the DiversityWorkbench is set (see below).
Info site tab. If unchanged, the default
site
for the DiversityWorkbench is set (see below).
If for any reason a user wants his name to be removed from the database,
select the users name from the list as shown below and click on
the  button (see below).
button (see below).
Diversity Collection
No-SQL Interface
JSON Cache
Databases of the modules provid a No-SQL interface where the data of the main table and the depending tables are merged as JSON.
Table JsonCache
Content of table JsonCache restricted to public available data
| Column |
Data type |
Description |
Nullable |
Relation |
| ID |
int |
Unique ID for the Dataset, Primary key |
NO |
ID of the source |
| URI |
varchar (500) |
The URL as combination of BaseURL and ID |
NO |
BaseURL and ID of the source |
| DisplayText |
nvarchar (500) |
Representation in the interface |
NO |
Main table of the source |
| LogUpdatedWhen |
datetime |
Date and time when the data were last updated |
NO |
- |
| Data |
json |
Data related to the current dataset |
NO |
- |
procFillJsonCache is started by an update trigger trgUpd… of the main table in the database
Interface in clients
All modules provide data via a cache table. In the header of the clients you can inspect the content of the JsonCache with a click on the  button. For modules with a difference between local and public data, you can inspect the content of the public data with a click of the right mouse button.
button. For modules with a difference between local and public data, you can inspect the content of the public data with a click of the right mouse button.
Update
Apart of the update via the Trigger (see below) you can update the JsonCache via the  update button underneath the
update button underneath the  button.
button.
To update the JsonCache for the whole database select Administration - JsonCache… from the menu. a window as shown below will open where you can update the JsonCache for single datasets or the whole database.
Summary
graph TD;
TaxonName[Main table in database]
trgUpdTaxonName[trgUpd.. of main table in database]
TaxonName --> |Update in table| trgUpdTaxonName
proc[Procedure procFillJsonCache setting the content in table JsonCache]
trgUpdTaxonName --> proc
graph TD;
Mainform[Main form]
ButtonShow[Button show JsonCache of current dataset]
Mainform --> ButtonShow
Left[Show Data]
ButtonShow --> |Left click| Left
graph TD;
Mainform[Main form]
Admin[Administration menu]
Mainform --> Admin
Cache[JsonCache...]
Admin --> Cache
Adminform[Administration form]
Cache --> Adminform
AdminUpdateSingle[Update single dataset]
Adminform --> AdminUpdateSingle
AdminUpdateDB[Update for whole database]
Adminform --> AdminUpdateDB
Example
The JsonCache of the current dataset will be shown as in the example below:
[
{
"Server": "http://zsm.diversityworkbench.de/",
"Database": "DiversityCollection_ZSM_Training",
"URL": "http://zsm.diversityworkbench.de/Collection_ZSM_Training/399050",
"Type": "CollectionSpecimen",
"ID": 399050,
"DisplayText": "JsonCacheDemo",
"Project": [
{
"Project": "HYMIScoll",
"ProjectID": 927
}
],
"CollectionEvent": [
{
"LocalityDescription": "Germany, Bavaria",
"CollectionYear": 2024,
"CollectionMonth": 7,
"CollectionDay": 9,
"CollectionEventLocalisation": [
{
"DisplayText": "Altitude (mNN)",
"Content Location 1": "Altitude from",
"Location1": "679",
"Content Location 2": "Altitude to"
},
{
"DisplayText": "Named area (DiversityGazetteers)",
"Content Location 1": "Location",
"Location1": "Holzleithen, Bavaria, Germany",
"Content Location 2": "Thesaurus code"
},
{
"DisplayText": "Coord. WGS84",
"Content Location 1": "Longitude (East-West)",
"Location1": "10.959893227",
"Content Location 2": "Latitude (North-South)",
"Location2": "47.753982544",
"Geography": "POINT (10.959893227 47.753982544)"
}
],
"CollectionEventProperty": [
{
"PropertyName": "Geographic regions",
"PropertyType": "Vegetation",
"Property": "Eschen-Ahorn-Schluchtwald bzw. -Hangwald (feucht-kühle Standorte)",
"Hierarchy": "Eschen-Ahorn-Schluchtwald bzw. -Hangwald (feucht-kühle Standorte) | Schlucht-, Blockhalden- und Hangschuttwälder | Laub(misch)wälder und -forste (Laubbaumanteil > 50 %)"
}
],
"CollectionEventMethod": [
{
"Method": "Fallen",
"Marker": "1",
"CCollectionEventParameterValue": [
{
"PropertyType": "Alkohol",
"Property": "70%"
},
{
"PropertyType": "Pheromon",
"Property": "-"
},
{
"PropertyType": "Licht",
"Property": "400 nm"
}
]
}
],
"CollectionEventRegulation": [
{
"Regulation": "DE_PER_20210801",
"TransactionTitle": "DE_PER_20210801"
}
],
"CollectionEventSeries": [
{
"SeriesID": -1,
"Description": "Voralpen",
"SeriesCode": "B2024_02",
"DateStart": "2024-07-01T13:29:22",
"DateEnd": "2024-07-27T13:29:22",
"CollectionEventSeriesDescriptor": [
{
"Descriptor": "Schluchtwälder",
"DescriptorType": "Biotoperfassung"
}
]
},
{
"SeriesID": -4,
"Description": "Ammerschlucht",
"Geography": "POINT (10.9668204567046 47.7016669695774)",
"DateStart": "2024-07-09T13:29:22",
"DateEnd": "2024-07-10T13:29:22",
"CollectionEventSeriesDescriptor": [
{
"Descriptor": "Schluchtwälder",
"DescriptorType": "Biotoperfassung"
}
]
}
],
"ExternalIdentifier": [
{
"Type": "LSID",
"Identifier": "Event01"
}
]
}
],
"Annotation": [
{
"Annotation": "Annotation for specimen"
}
],
"CollectorsName": [
{
"CollectorsName": "Huber, F.",
"CollectorsNumber": "FH-654"
},
{
"CollectorsName": "Pfeiffer, R."
},
{
"CollectorsName": "Schmidt, H."
}
],
"CollectionSpecimenReference": [
{
"ReferenceTitle": "Reference for a part",
"SpecimenPartID_Reference": 182805
}
],
"CollectionSpecimenRelation": [
{
"RelatedSpecimenDisplayText": "ext. Relation of specimen to something else",
"RelationType": "Specimen part",
"RelatedSpecimenURI": "ext. Relation of specimen to something else"
}
],
"CollectionSpecimenPart": [
{
"SpecimenPartID": 182807,
"MaterialCategory": "DNA sample",
"CollectionHierarchy": [
{
"CollectionName": "Entomologie",
"CollectionAcronym": "Ento",
"Type": "department"
},
{
"CollectionName": "Insecta varia",
"CollectionAcronym": "InVa",
"CollectionOwner": "SNSB - Zoologische Staatssammlung München",
"Type": "department"
},
{
"CollectionName": "Odonata",
"Type": "department"
},
{
"CollectionName": "SNSB - Zoologische Staatssammlung München",
"CollectionAcronym": "ZSM",
"Type": "institution"
}
],
"CollectionSpecimenPartProcessing": [
{
"DisplayText": "DNA",
"ProcessingDate": "2024-07-10T10:47:53.050",
"ResponsibleName": ""
}
],
"CollectionSpecimenTransaction": [
{
"TransactionType": "loan",
"TransactionTitle": "Ausleihe nach berlin",
"CollectionName": "SNSB - Zoologische Staatssammlung München",
"ActualEndDate": "2024-07-04T00:00:00"
}
],
"CollectionSpecimenTask": [
{
"Task": "Exhibition ⁞ Part",
"Type": "Part",
"CollectionName": "Ebene 2",
"DisplayText": "JsonCacheDemo: Calopteryx virgo(Linnaeus, 1758) - DNA sample"
}
]
},
{
"SpecimenPartID": 182805,
"MaterialCategory": "pinned specimen",
"CollectionHierarchy": [
{
"CollectionName": "ZSM-InVa-D1482",
"CollectionAcronym": "10",
"Type": "drawer",
"CollectionImage": [
{
"URI": "https://www.zsm.mwn.de/drawers/InVa/Odonata/ZSM-InVa-D1482-2020"
}
]
}
],
"CollectionSpecimenTask": [
{
"Task": "Exhibition ⁞ Part",
"Type": "Part",
"CollectionName": "Ebene 2",
"DisplayText": "JsonCacheDemo: Calopteryx virgo(Linnaeus, 1758) - pinned specimen"
}
]
},
{
"SpecimenPartID": 182806,
"MaterialCategory": "tissue sample",
"CollectionHierarchy": [
{
"CollectionName": "ZSM-InVa-D1482",
"CollectionAcronym": "10",
"Type": "drawer",
"CollectionImage": [
{
"URI": "https://www.zsm.mwn.de/drawers/InVa/Odonata/ZSM-InVa-D1482-2020"
}
]
}
],
"CollectionSpecimenTransaction": [
{
"TransactionType": "regulation",
"TransactionTitle": "DE_PER_20210801",
"CollectionName": "SNSB - Zoologische Staatssammlung München"
}
],
"CollectionSpecimenTask": [
{
"Task": "Exhibition ⁞ Part",
"Type": "Part",
"CollectionName": "Ebene 2",
"DisplayText": "JsonCacheDemo: Calopteryx virgo(Linnaeus, 1758) - tissue sample"
}
]
}
],
"IdentificationUnit": [
{
"IdentificationUnitID": 437340,
"TaxonomicGroup": "fungus",
"Identification": [
{
"TaxonomicName": "Metarhizium anisopliae",
"IdentificationSequence": 0
}
]
},
{
"IdentificationUnitID": 437339,
"HierarchyCache": "Odonata - Calopterygidae - Calopteryginae",
"OrderCache": "Odonata",
"FamilyCache": "Calopterygidae",
"TaxonomicGroup": "insect",
"NumberOfUnits": 22,
"Identification": [
{
"TaxonomicName": "Calopteryx virgo(Linnaeus, 1758)",
"IdentificationSequence": 1
}
],
"IdentificationUnitAnalysis": [
{
"DisplayText": "16S",
"AnalysisNumber": "1",
"AnalysisResult": "TGC",
"SpecimenPartID": 182805
},
{
"DisplayText": "DNA Analysis",
"AnalysisDate": "2024-06-24",
"AnalysisNumber": "1",
"AnalysisResult": "ATTGCAGC",
"ResponsibleName": "Meier, R.",
"IdentificationUnitAnalysisMethod": [
{
"DisplayText": "Ultraschall",
"MethodMarker": "1"
}
]
}
],
"IdentificationUnitGeoAnalysis": [
{
"AnalysisDate": "2024-07-08T14:39:21.300",
"Geography": "POINT (11.50062829 48.16385096)"
},
{
"AnalysisDate": "2024-07-08T14:45:28.007",
"Geography": "POLYGON ((11.5003972353666 48.1634295515789, 11.5005823309247 48.1634194298878, 11.5005903785577 48.1634788947945, 11.5004012591831 48.1635041989893, 11.5003972353666 48.1634295515789))"
}
]
}
]
}
]
Cache database
Table [Project].CacheJsonCache
Content of table JsonCache restricted to public available data in the Cache database
| Column |
Data type |
Description |
| ID |
int |
Unique ID for the Dataset, Primary key |
| URI |
varchar (500) |
The URL as combination of BaseURL and ID |
| DisplayText |
nvarchar (500) |
Representation in the interface |
| LogUpdatedWhen |
datetime |
Date and time when the data were last updated |
| Data |
json |
Public available data related to the current dataset |
The data are transferred via a stored procedure [Project].procPublishJsonCache where [Project] is the schema corresponding to the Project in the database. Restricted to public information e.g. not locked via DataWithholding, Internal…, etc. and further restrictions as defined in the cache database.
Postgres database
The table CacheJsonCache is a copy of the table in the SQL-Server database with the Data stored in JSONB Format (= binary).
 Series
Series collection event or observations at single spots
collection event or observations at single spots Event
Event a specimen was collected during, for example, an
a specimen was collected during, for example, an  expedition
expedition Localisation
Localisation Site Property
Site Property Collecting Method
Collecting Method Specimen
Specimen Collector
Collector Objects
Objects Identification
Identification Analysis
Analysis Analysis methods
Analysis methods Part
Part Collection
Collection Processing
Processing Processing methods
Processing methods Transaction
Transaction Project
Project Images
Images References
Referencesmay have been collected at a CollectionEvent
during an expedition.
. The site of the CollectionEvent can be localised
and characterised
and methods
have been used. The collectors
collected twigs of the plant
and fungi
 growing on the plants.
growing on the plants.several times and analysed
where methods
have been used.
 and specimens
and specimens
in a collection.
.
 sent
some of the samples
sent
some of the samples for which he had a request
 from a requester
from a requester
to another collection.
.
 and DNA
and DNA  extracted which was processed
extracted which was processed where methods
have been used.
were taken for the EventSeries, the CollectionEvent, the CollectionSpecimen as well as organisms and parts of this specimen.
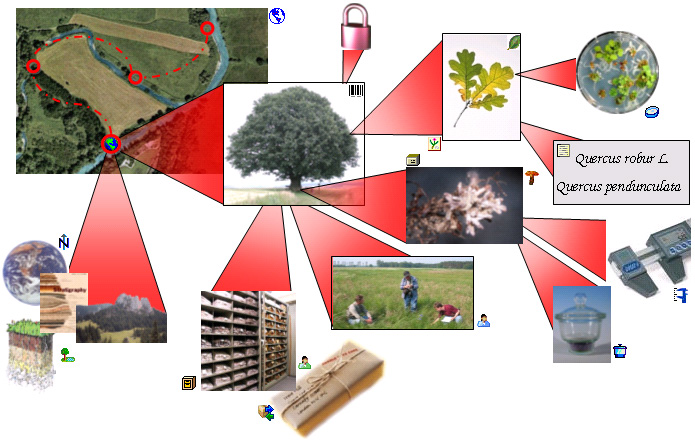
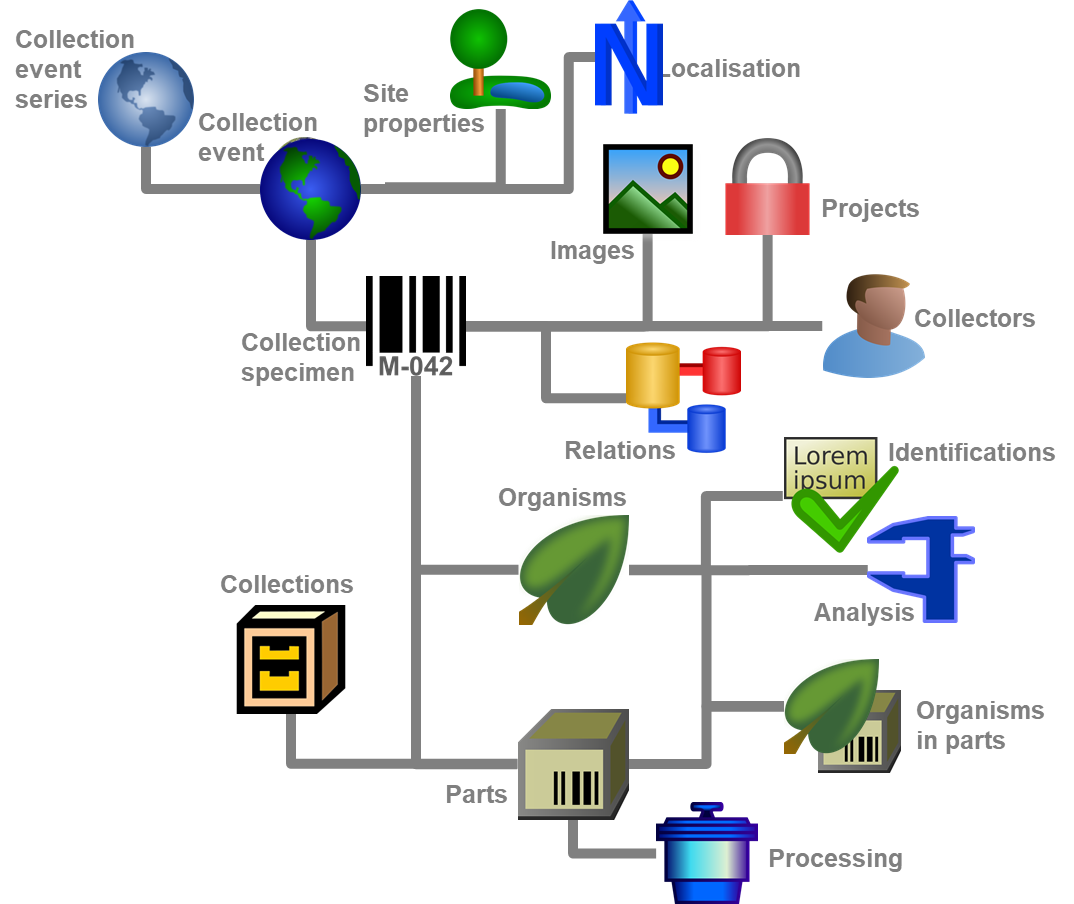



 Administration: Database: Create new login: Fix link to DiversityAgents
Administration: Database: Create new login: Fix link to DiversityAgents  ImportWizard: Optimizing startup of
ImportWizard: Optimizing startup of 
 Enabling
Enabling  translation .
translation .  New Property DerivedFromInternalParentRelation for datacolumns to import IDs in columns related to the PK of the table.
New Property DerivedFromInternalParentRelation for datacolumns to import IDs in columns related to the PK of the table.  Message for wrong encoding improved.
Message for wrong encoding improved. Translation of old encodings into new nomenclature.
Translation of old encodings into new nomenclature. Plans in collection
Plans in collection
 Expand zoom to minimal 1% of size to ensure visibility of whole plan.
Expand zoom to minimal 1% of size to ensure visibility of whole plan. Show geometries of child collections.
Show geometries of child collections.
 Show names of child collections.
Show names of child collections. CacheDB -
CacheDB -  Package FloraRaster -
Package FloraRaster -  DiversityCollectionUpdate_020656 - DiversityCollectionUpdate_020657
DiversityCollectionUpdate_020656 - DiversityCollectionUpdate_020657
 Transfer selected data to
Transfer selected data to 
 Link selected data to
Link selected data to 


 Administration - Collections: Redesign of
Administration - Collections: Redesign of  Administration - Collections: Refactor of hierarchy calculation via
Administration - Collections: Refactor of hierarchy calculation via  Refactoring
Refactoring  New taxonomic group ‘organism’, as ‘TaxonomyRelatedTaxonomicGroups’ (fields Family and Order).
New taxonomic group ‘organism’, as ‘TaxonomyRelatedTaxonomicGroups’ (fields Family and Order). New material category core sample
New material category core sample Labelprinting: Fixed a problem when creating labels with author names starting with ‘de’.
Labelprinting: Fixed a problem when creating labels with author names starting with ‘de’. Datasets:
Datasets: Default for timeout for loading resources from web (e.g. images) set to 1 sec: TimeoutWeb in DiversityWorkbench.WorkbenchSettings.settings - TimeoutWeb.
Default for timeout for loading resources from web (e.g. images) set to 1 sec: TimeoutWeb in DiversityWorkbench.WorkbenchSettings.settings - TimeoutWeb.  Inclusion of request.
Inclusion of request. descriptor icon in query.
descriptor icon in query. Maintenance
Maintenance
 Exhibition including option for printing
Exhibition including option for printing Collection: Transfer to parent location based on location hierarchy
Collection: Transfer to parent location based on location hierarchy

 FirstLines_4: Optimized Query for IdentificationUnit
FirstLines_4: Optimized Query for IdentificationUnit Granting delete and update for Annotation to editor
Granting delete and update for Annotation to editor Disable triggers for update of related tables Annotation and ExternalIdentifier
Disable triggers for update of related tables Annotation and ExternalIdentifier Statistics
Statistics
 Inclusion of optional setting for charts for queries linked to modules
Inclusion of optional setting for charts for queries linked to modules IPM: Redesign of collection hierarchy
IPM: Redesign of collection hierarchy Setting of user defined service for QR-Code generation.
Setting of user defined service for QR-Code generation. ExternalIdentifier
ExternalIdentifier
 Datawithholding reason for specimen shown in label and tooltip
Datawithholding reason for specimen shown in label and tooltip Export
Export
 Bavarikon:
Bavarikon:
 Balance:
Balance:  Redesign of handling regulations:
Redesign of handling regulations:

 Descriptors for the event series
Descriptors for the event series  Customize: Option for display of valid agent name included
Customize: Option for display of valid agent name included disable IX_CollectionAgentSequence if present to ensure insert
disable IX_CollectionAgentSequence if present to ensure insert Mainform - hierarchy nodes
Mainform - hierarchy nodes Spreadsheet:
Spreadsheet: TK-sheet includes hidden RowGUID for collectors by default
TK-sheet includes hidden RowGUID for collectors by default Documentation
Documentation
 Archive
Archive
 Optional inclusion of log tables
Optional inclusion of log tables HUGO
HUGO
 Updating depending datasets
Updating depending datasets  Adding current dataset.
Adding current dataset. Adding all datasets from the list.
Adding all datasets from the list. Showing content of the list.
Showing content of the list. Clearing the list.
Clearing the list. Showing message for button ShowDetail if no detail is defined
Showing message for button ShowDetail if no detail is defined
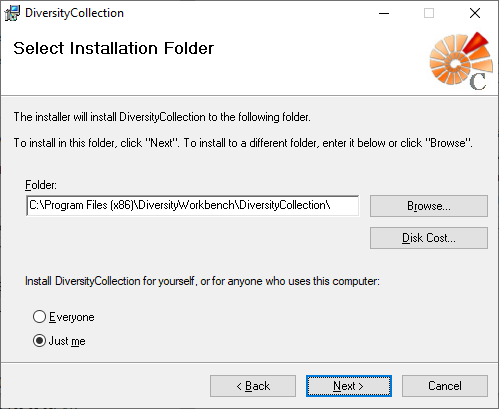

 . A dialog form “Connect to database” opens.
. A dialog form “Connect to database” opens.
 .
. .
.
 next to the Connect to server button indicates an encrypted connection.
By clicking on the icon, you can switch to an unencrypted connection, indicated by the icon
next to the Connect to server button indicates an encrypted connection.
By clicking on the icon, you can switch to an unencrypted connection, indicated by the icon  .
.



 Current acivity on server: List the processes on the database server.
Current acivity on server: List the processes on the database server. Ask on exit: The application should confirm the exit if the main window is closed.
Ask on exit: The application should confirm the exit if the main window is closed. Quit: Quit the application and stop all processes started by the application.
Quit: Quit the application and stop all processes started by the application.
 Event parameter table …: Edit the values of the parameters for the methods used in a collection event.
Event parameter table …: Edit the values of the parameters for the methods used in a collection event. Collector table …: Edit data of the collector table.
Collector table …: Edit data of the collector table. Relation table …: Edit data of the specimen relation table.
Relation table …: Edit data of the specimen relation table. Geo analysis table …: Edit data in the geo analysis table.
Geo analysis table …: Edit data in the geo analysis table. Part description …: Edit data of the specimen part description table.
Part description …: Edit data of the specimen part description table. Organism in part …: Edit data of the organism in part description table.
Organism in part …: Edit data of the organism in part description table. Specimen transaction …: Edit data of the collection specimen transaction table.
Specimen transaction …: Edit data of the collection specimen transaction table. Mineral sheet …: Edit mineral and related data with one line per mineral.
Mineral sheet …: Edit mineral and related data with one line per mineral. Scan mode: Change to scan mode to open the data set of a specimen by scanning the barcode.
Scan mode: Change to scan mode to open the data set of a specimen by scanning the barcode. Scan mode collection: Change to the collection scan mode to list all specimens within a collection by scanning the barcode that identifies that collection.
Scan mode collection: Change to the collection scan mode to list all specimens within a collection by scanning the barcode that identifies that collection.
 Import Observations …: Import observation data from tab-separated text files.
Import Observations …: Import observation data from tab-separated text files.



 Add publisher …: Add a replication publisher for the replication.
Add publisher …: Add a replication publisher for the replication. Download …: Download data from the data provider in your local database.
Download …: Download data from the data provider in your local database. Merge …: Merge contents between your local database and the replication provider.
Merge …: Merge contents between your local database and the replication provider. Upload …: Upload data from your local database to the replication provider.
Upload …: Upload data from your local database to the replication provider. Remove …: Remove the replication provider.
Remove …: Remove the replication provider. Clean database …: Clean your local database.
Clean database …: Clean your local database. Create archive …: Create and archive of project data.
Create archive …: Create and archive of project data. Restore archive …: Restore project data from an archive.
Restore archive …: Restore project data from an archive. Create schema …: Generate schema files describing the database tables for the project.
Create schema …: Generate schema files describing the database tables for the project.

 Save data set: Save current data set.
Save data set: Save current data set. Change password: Changing the password of a user (only available for SQL-Server accounts).
Change password: Changing the password of a user (only available for SQL-Server accounts).











 Expired loans …: Administration of expired loans. This menu entry will appear when expired loans are in collections where the current user is a curator.
Expired loans …: Administration of expired loans. This menu entry will appear when expired loans are in collections where the current user is a curator. My requests …: Administration of the loan requests of a user. This menu entry will appear when a user places requests for specimens.
My requests …: Administration of the loan requests of a user. This menu entry will appear when a user places requests for specimens. Collection users …: Administration of user access to data within collections.
Collection users …: Administration of user access to data within collections.
 Manual: Opens the online manual.
Manual: Opens the online manual.
 Feedback history …: Opens a window for browsing former feedback.
Feedback history …: Opens a window for browsing former feedback. Edit feedback …: Opens a window for editing feedback sent to the administrator (for admins only).
Edit feedback …: Opens a window for editing feedback sent to the administrator (for admins only).
 Websites: Websites related to DiversityCollection.
Websites: Websites related to DiversityCollection.
 Diversity mobile …: Website of DiversityMobile, the mobile application for collecting data stored in DiversityCollection.
Diversity mobile …: Website of DiversityMobile, the mobile application for collecting data stored in DiversityCollection. GitHub: Resouces on GitHub.
GitHub: Resouces on GitHub.






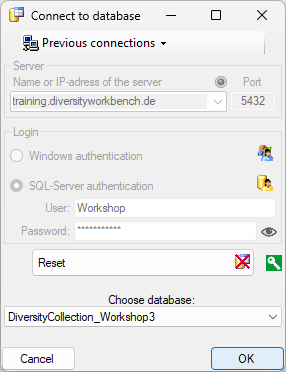

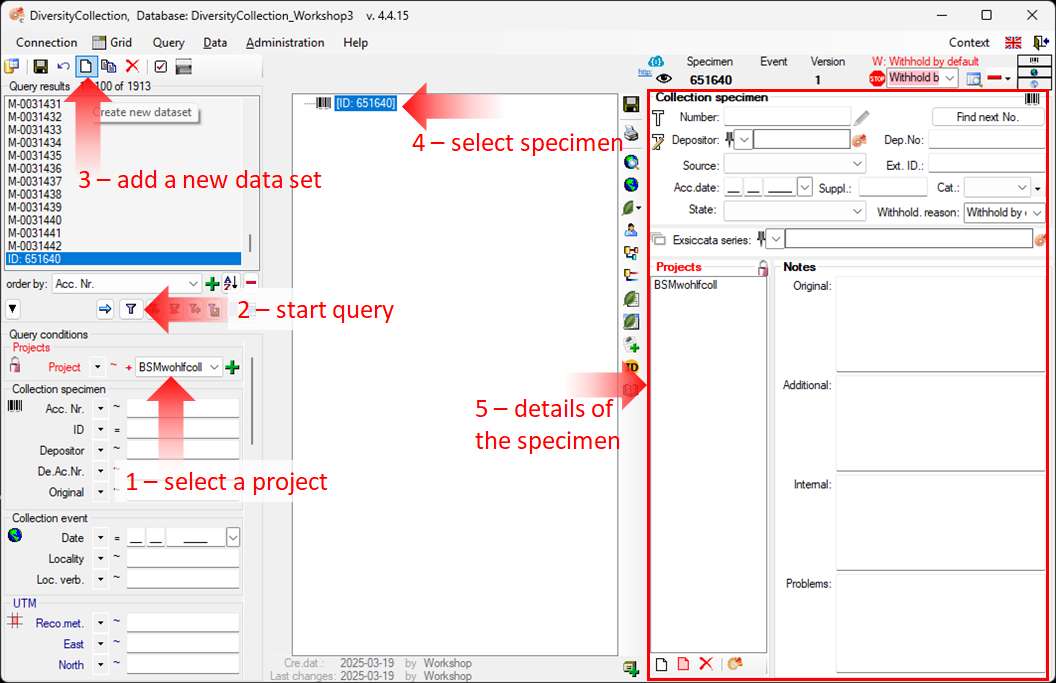
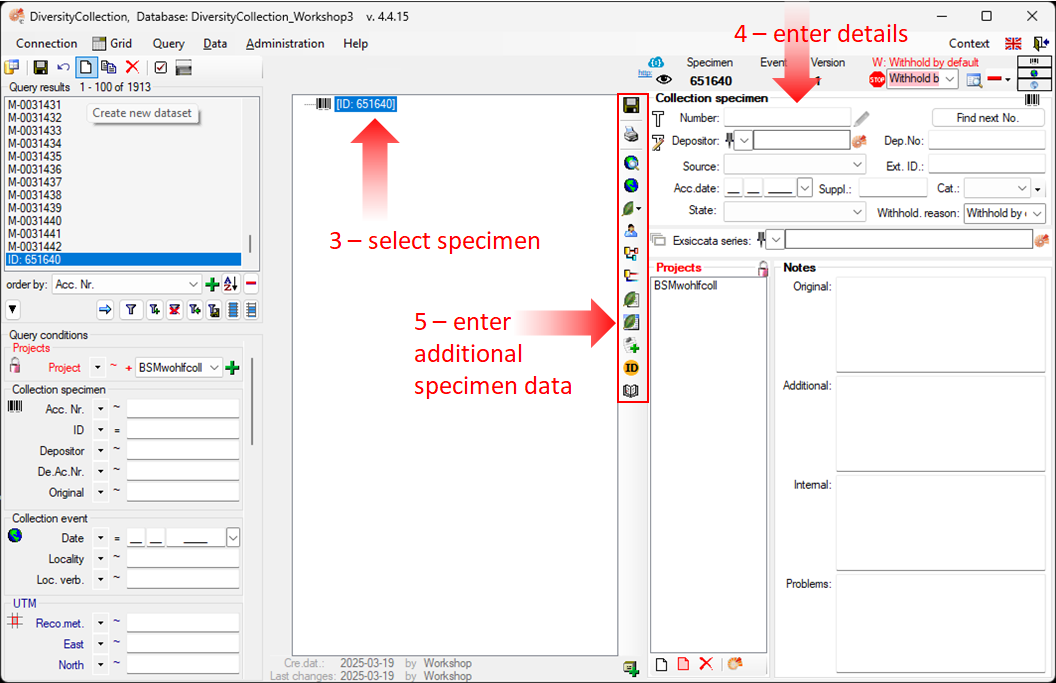
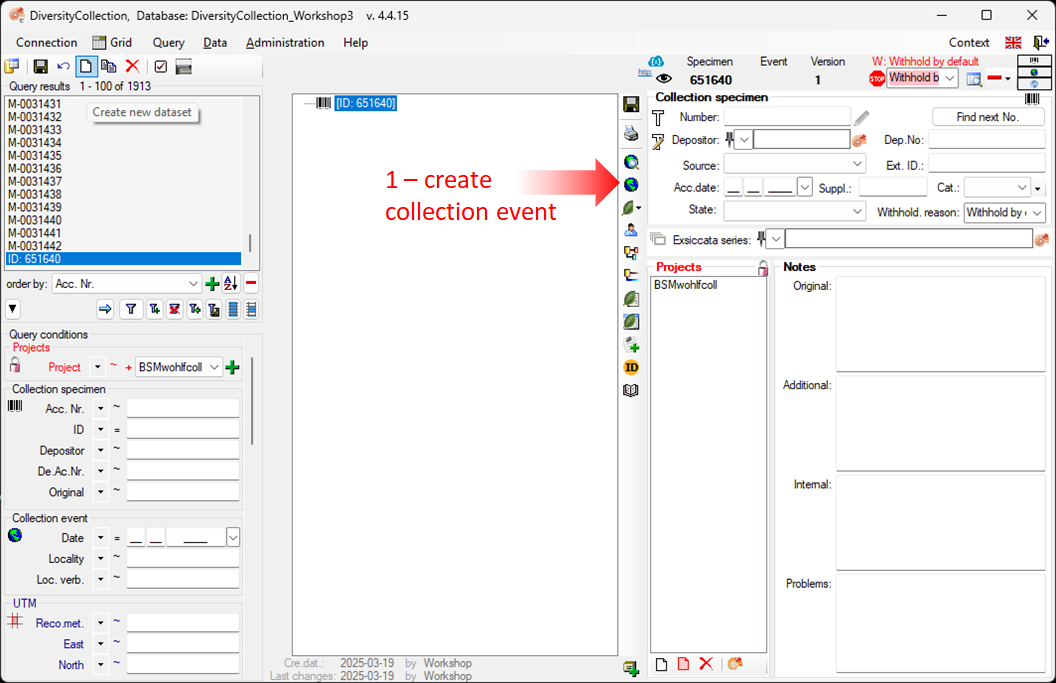
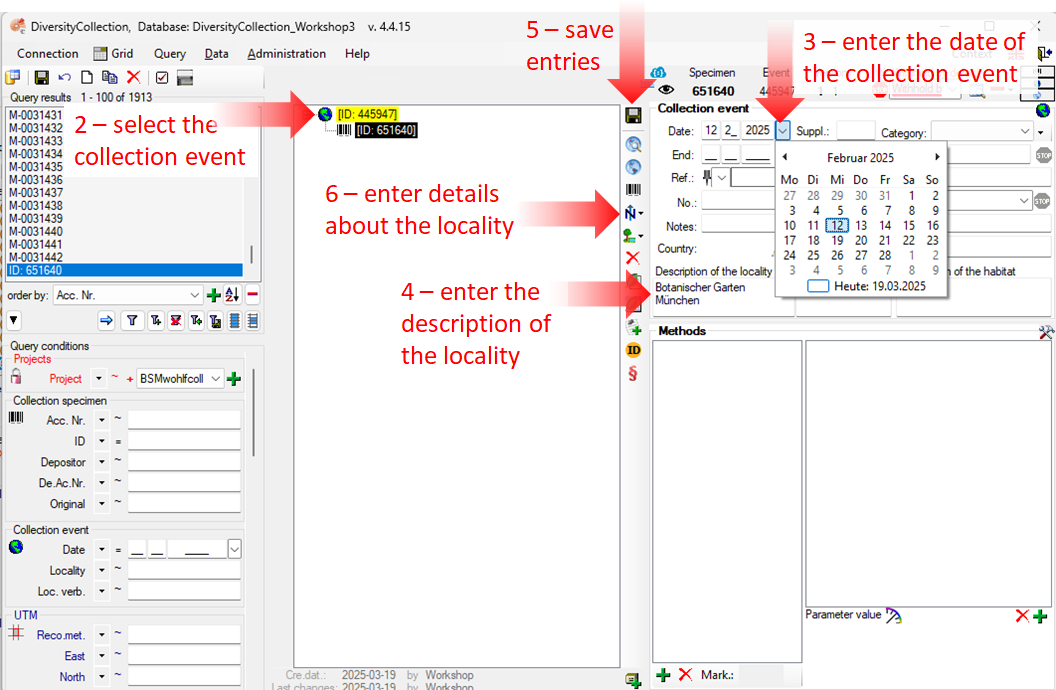
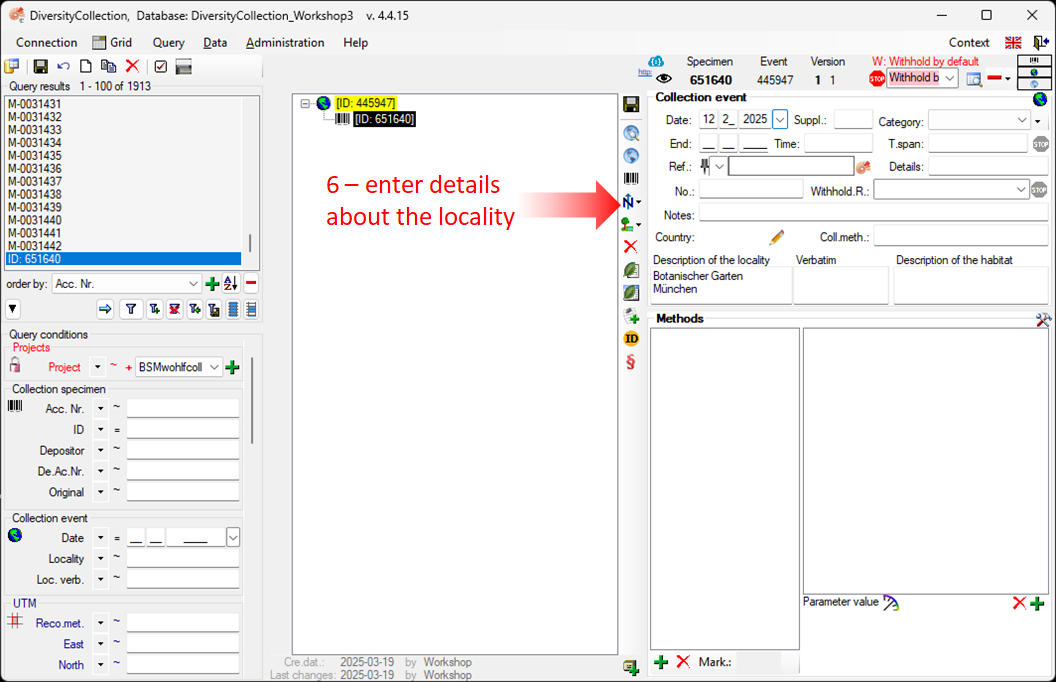
 (see point 2 in the image below).
(see point 2 in the image below).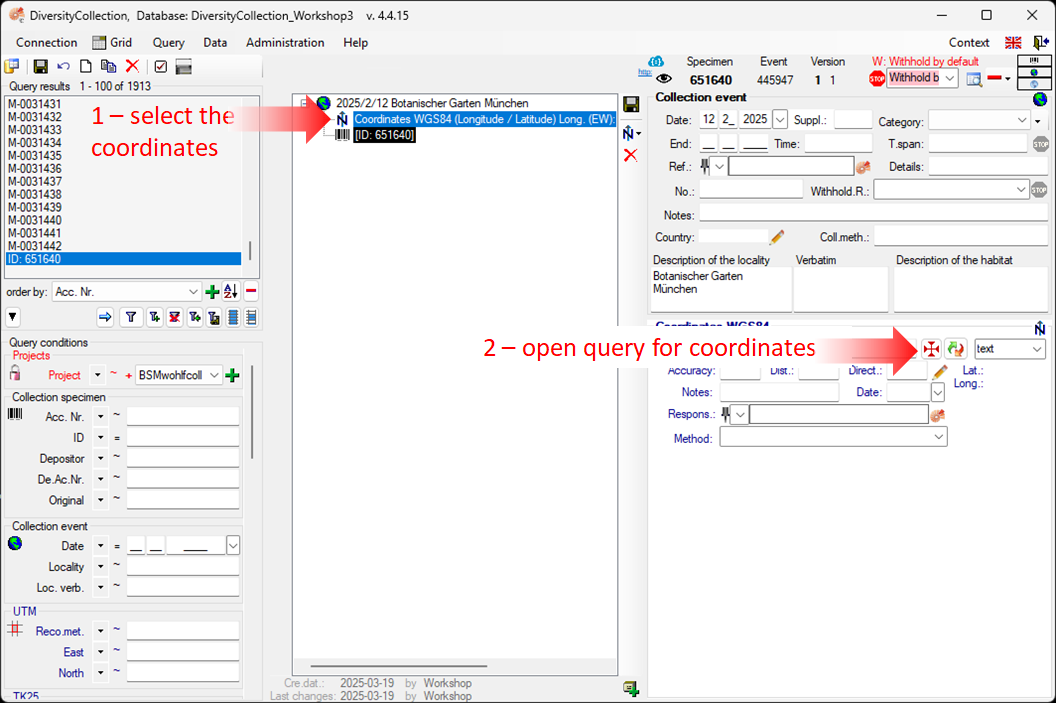
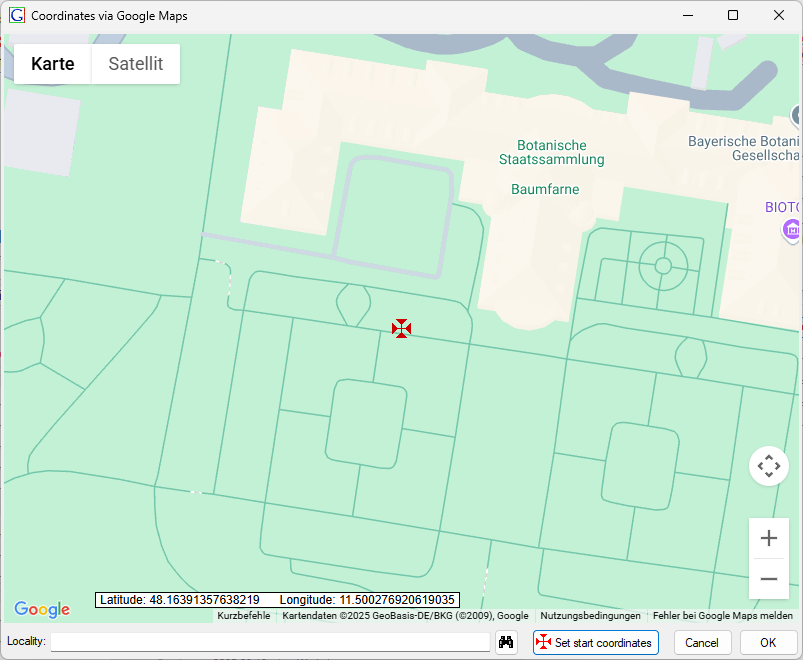
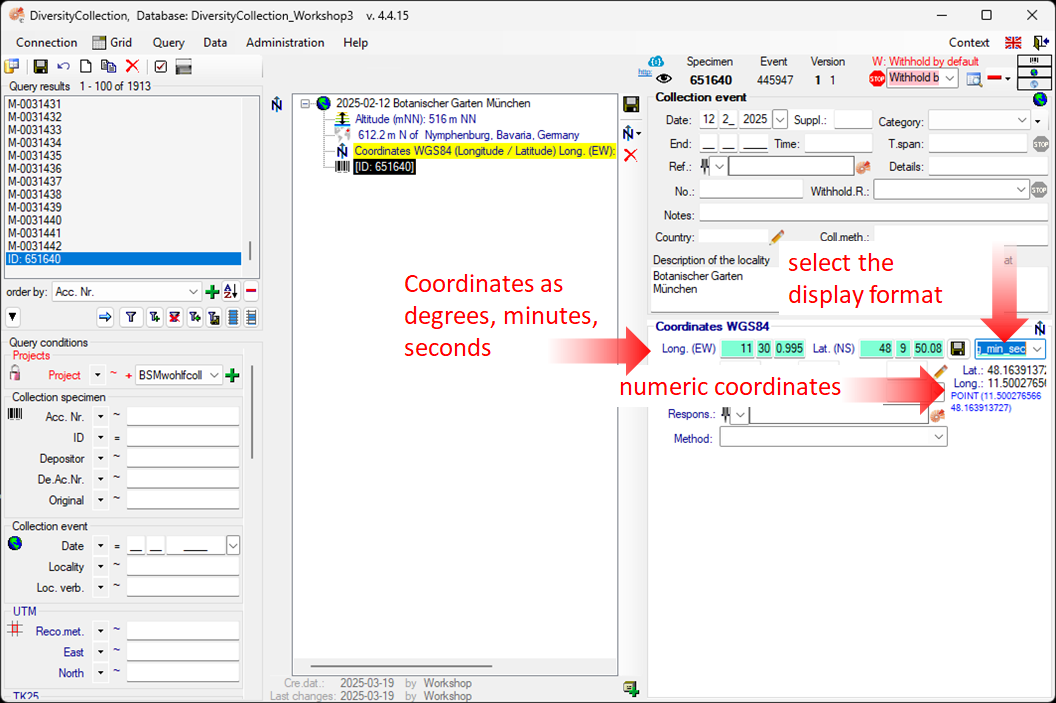
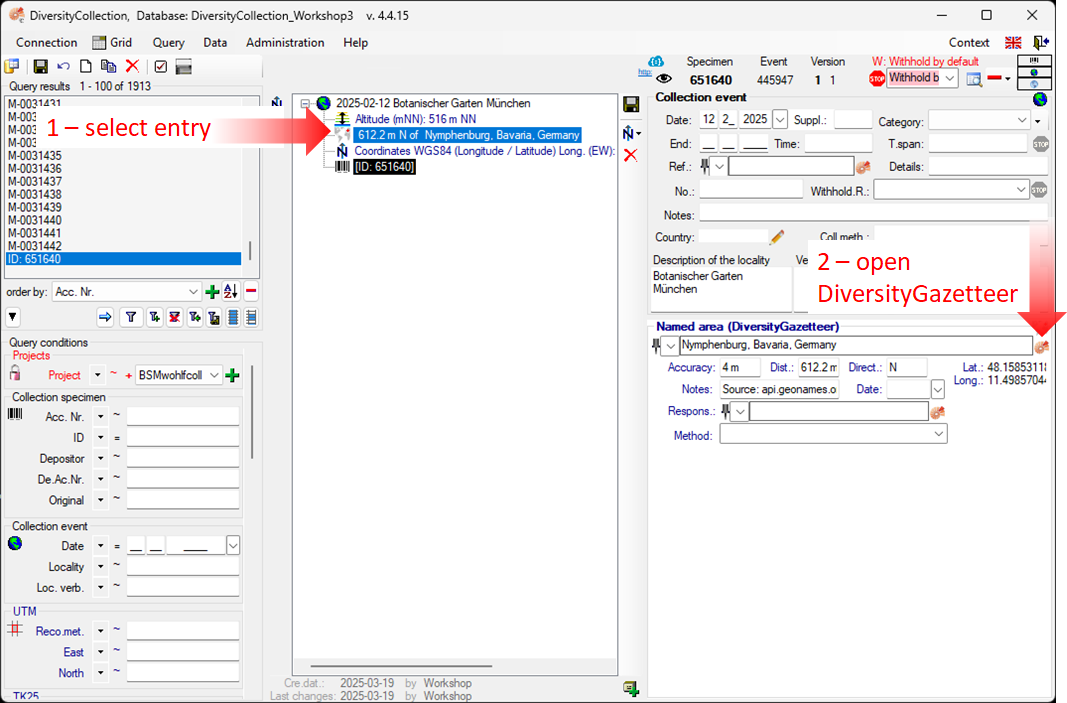
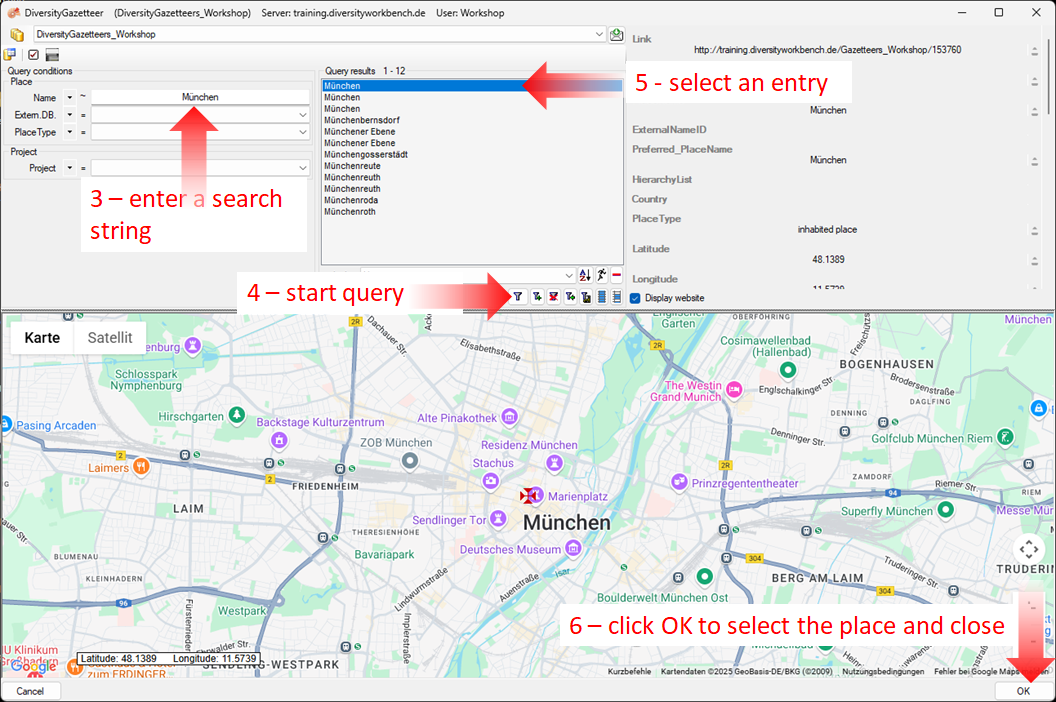
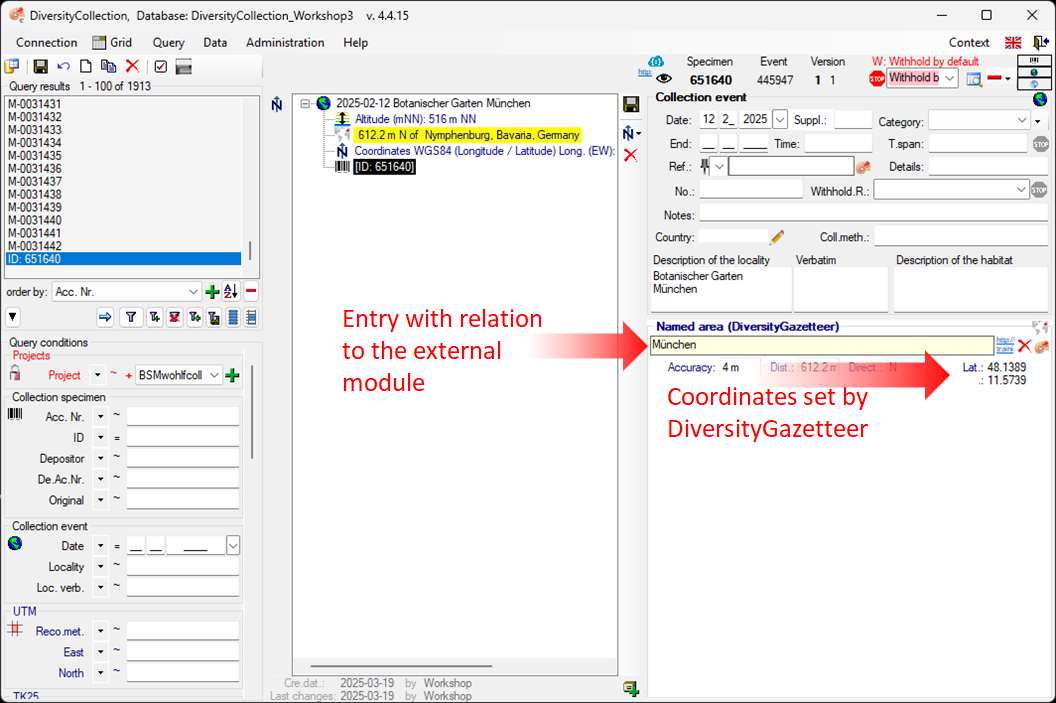
 the external module.
Click it for more details.
the external module.
Click it for more details.
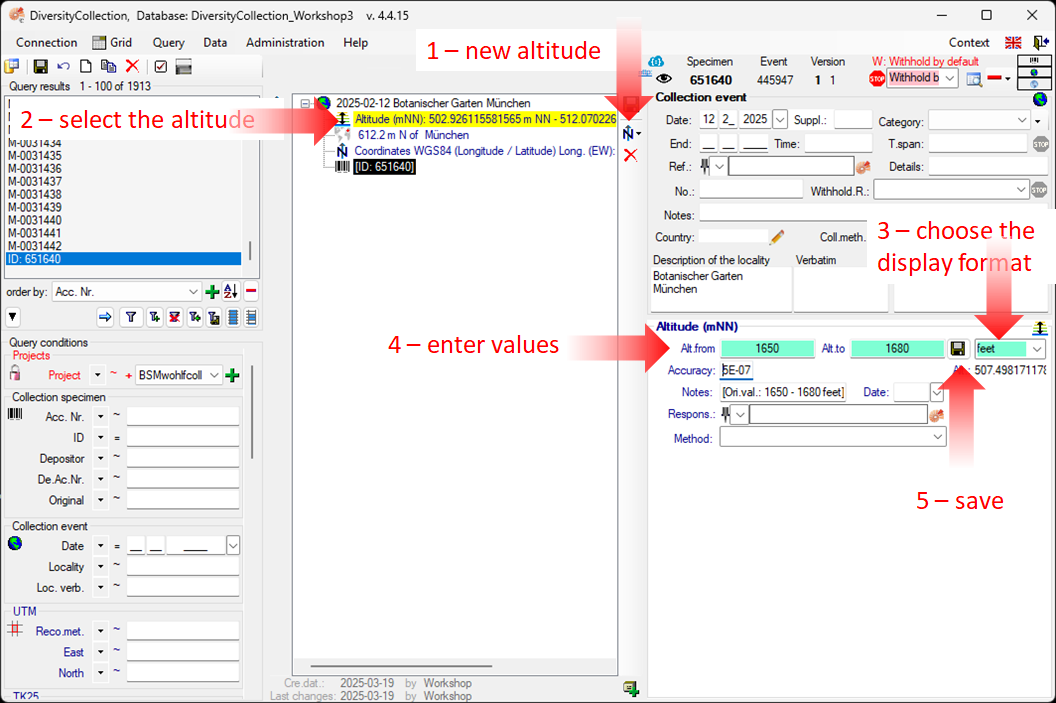
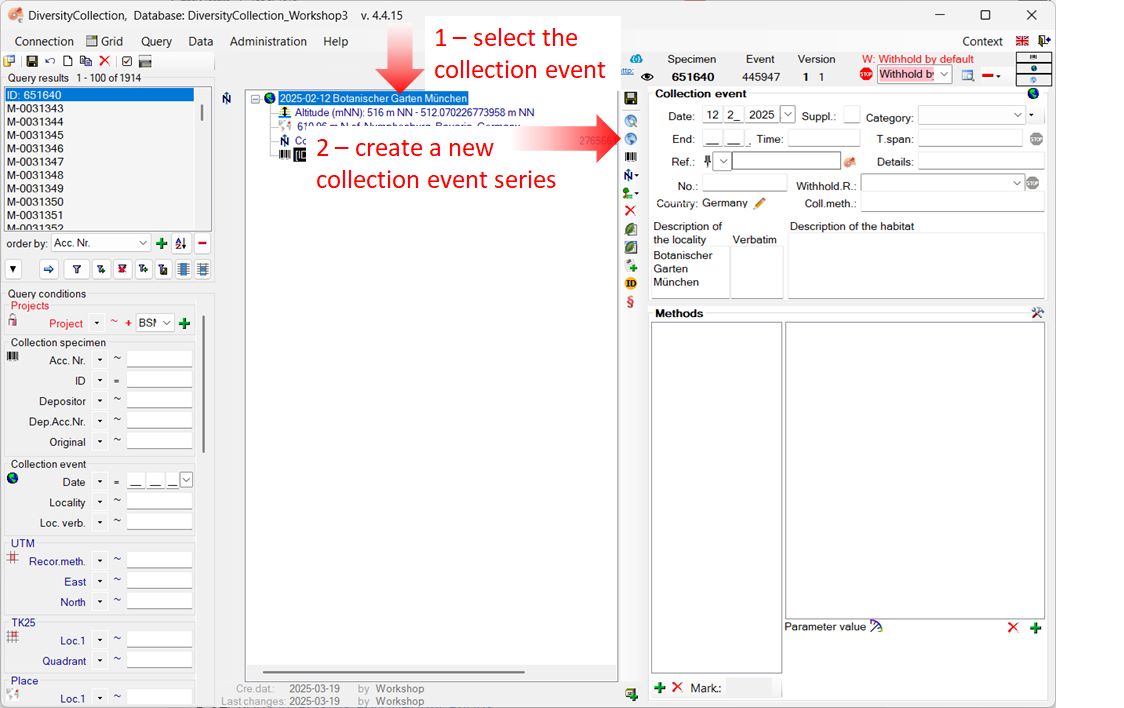
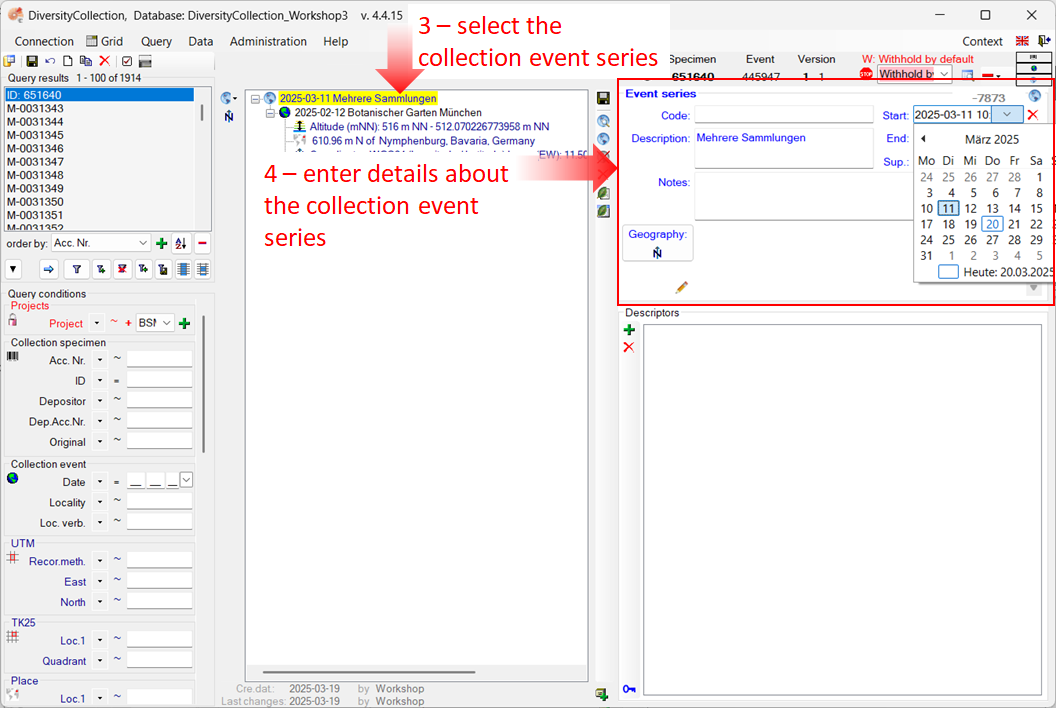
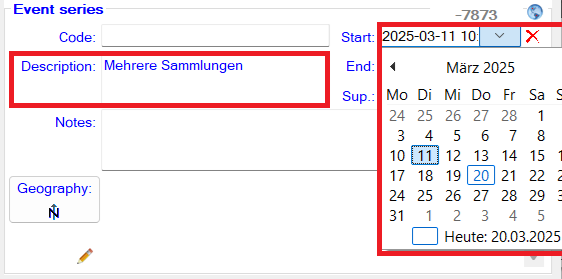
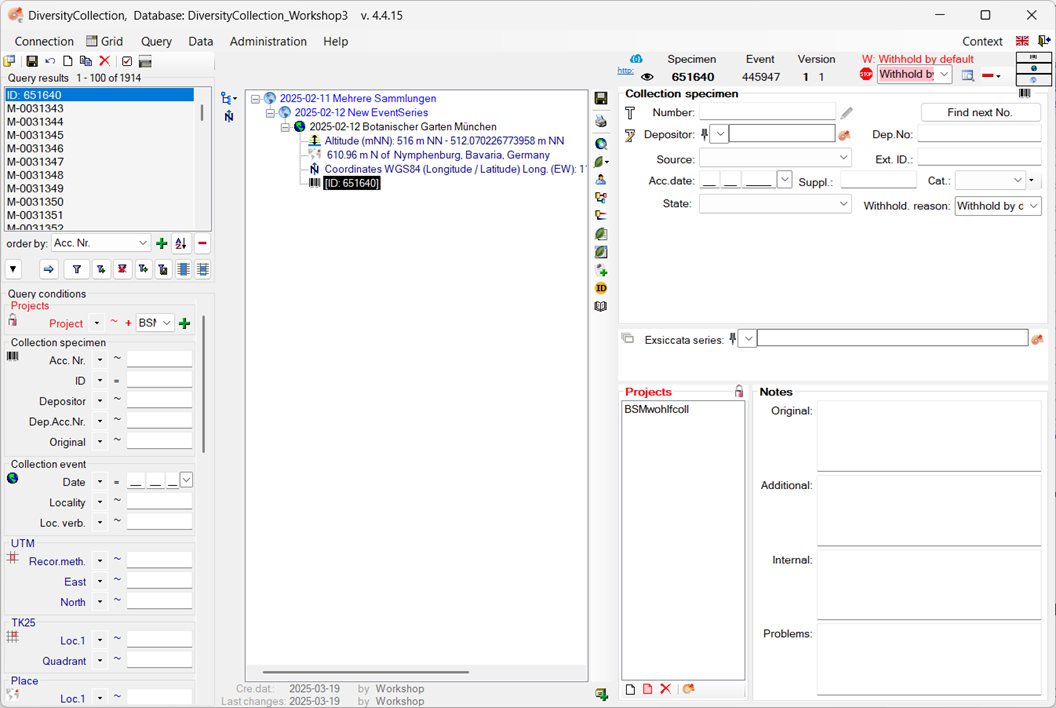
 .
.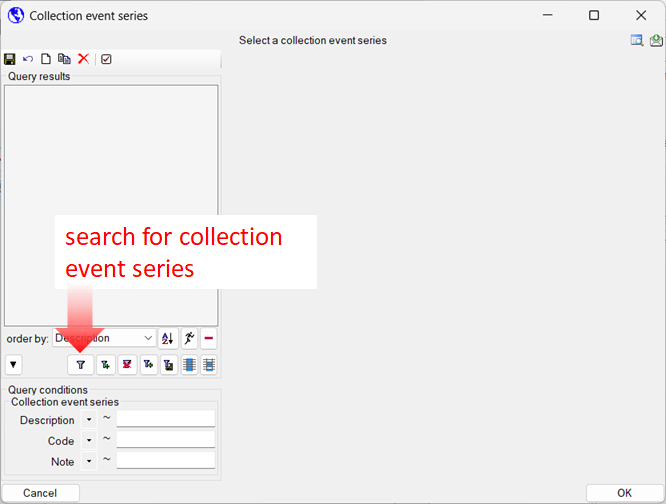
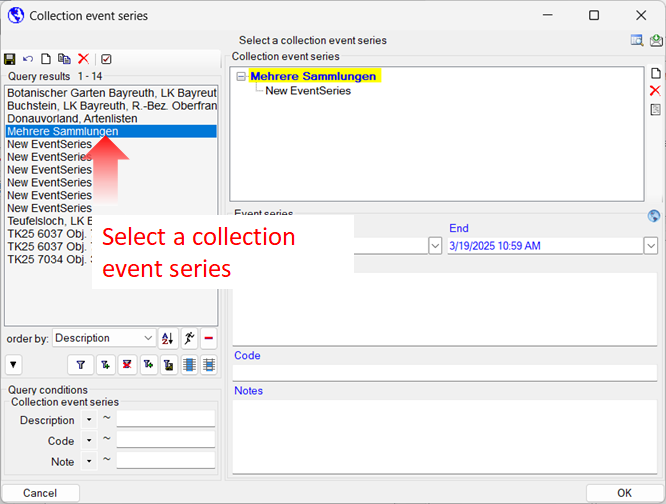
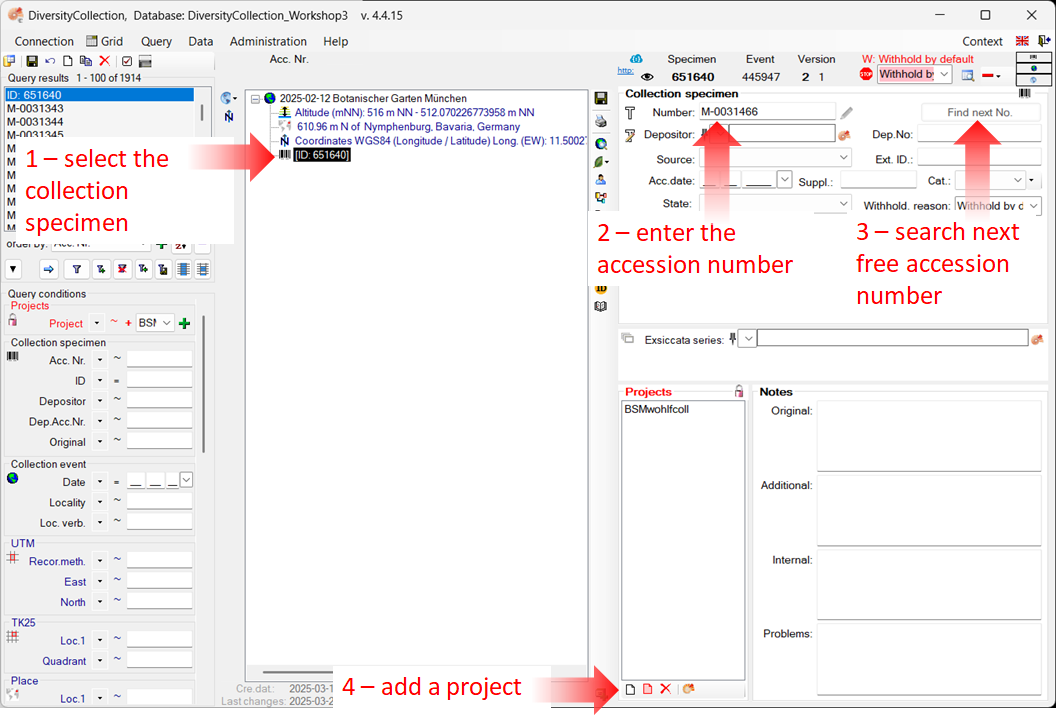
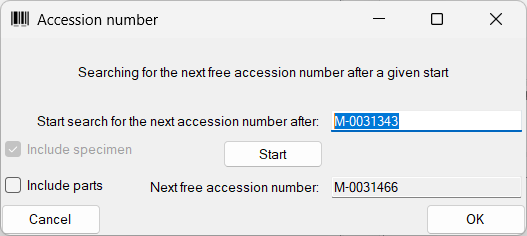
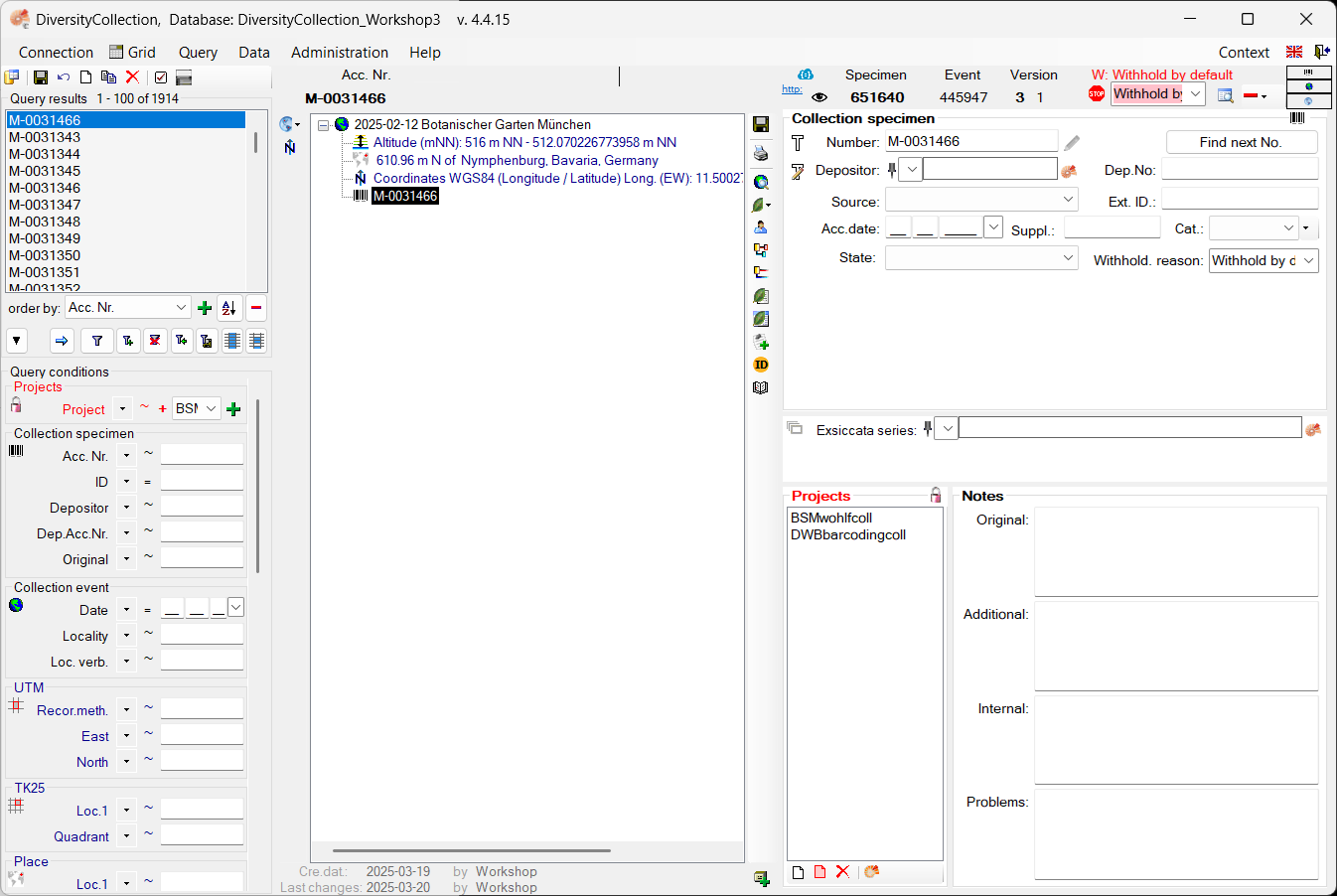
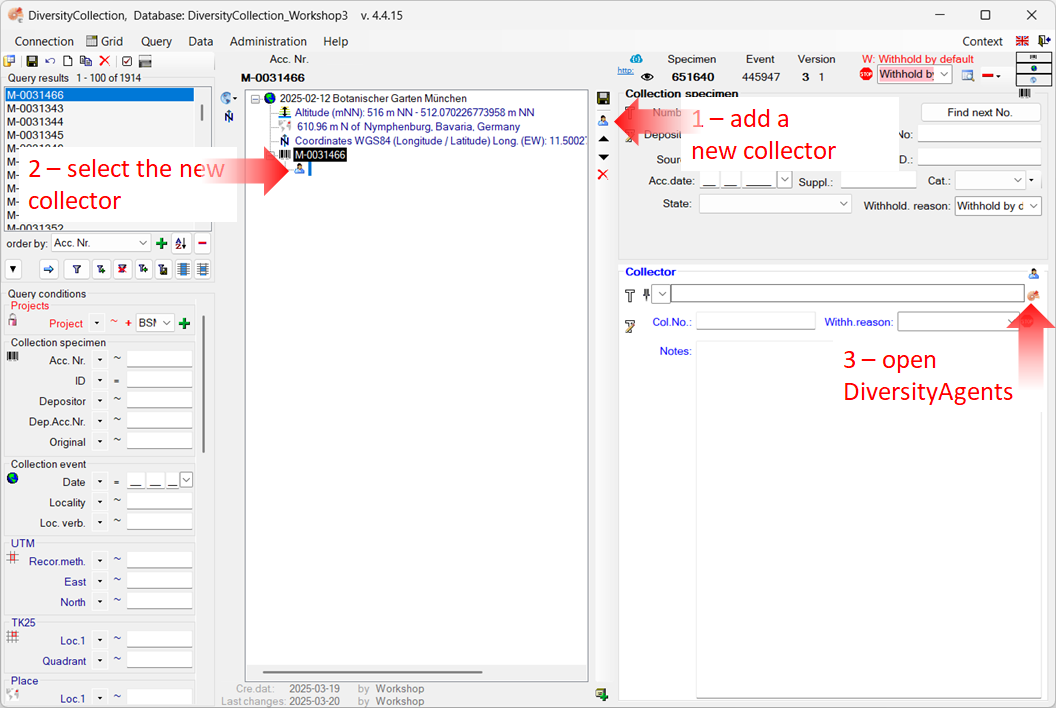
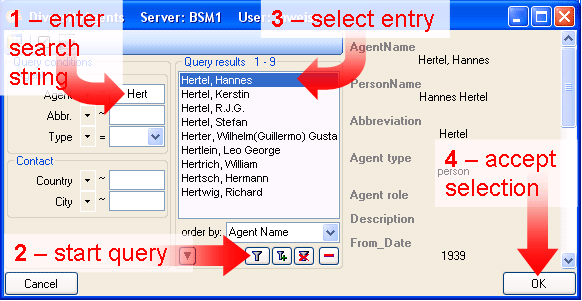
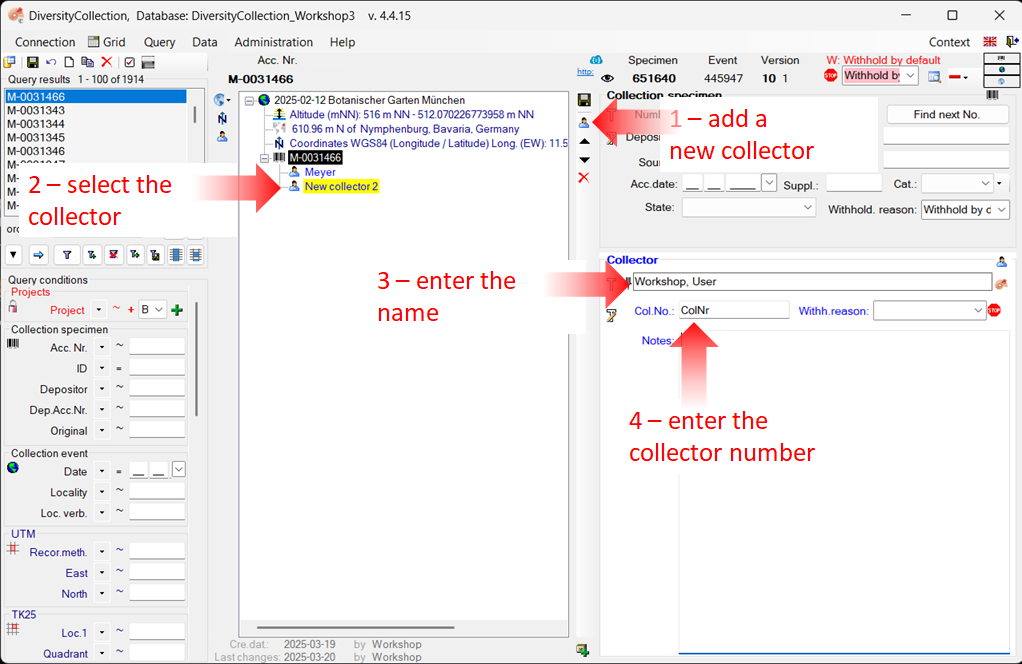
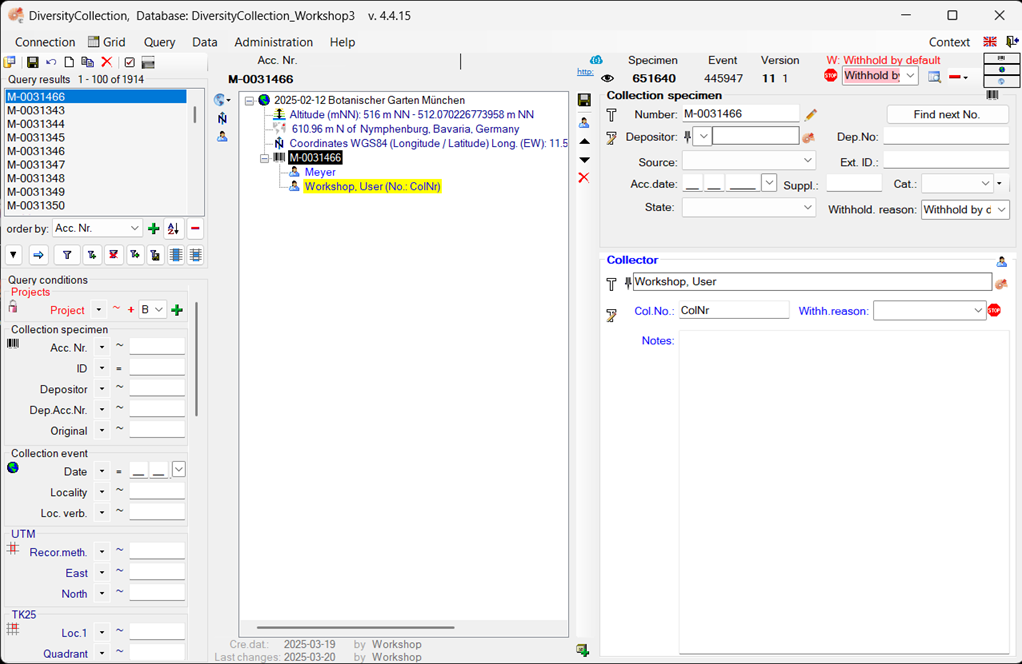

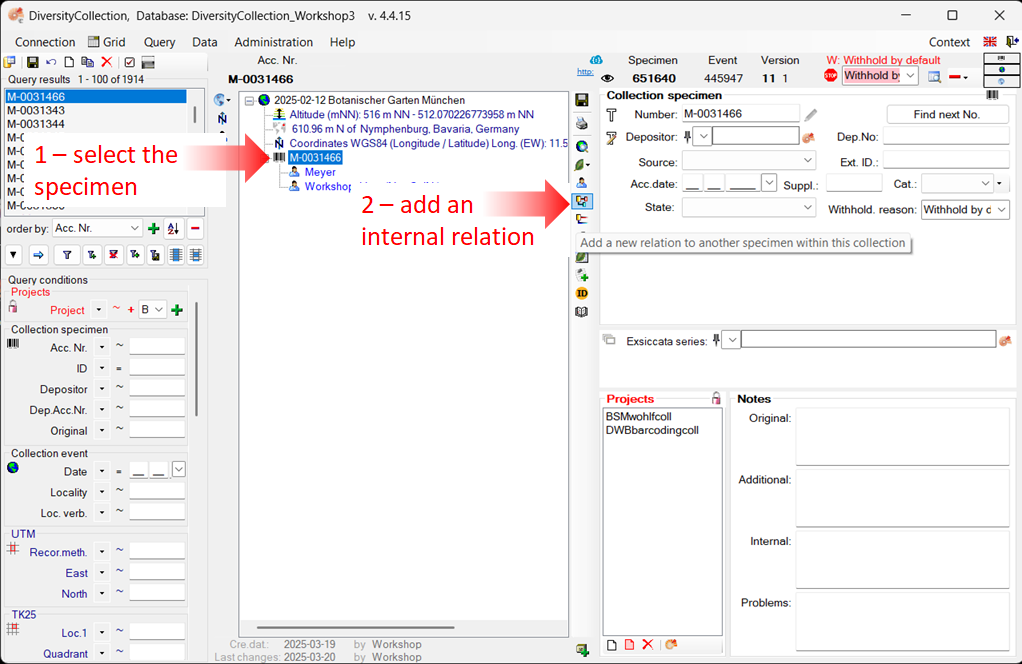
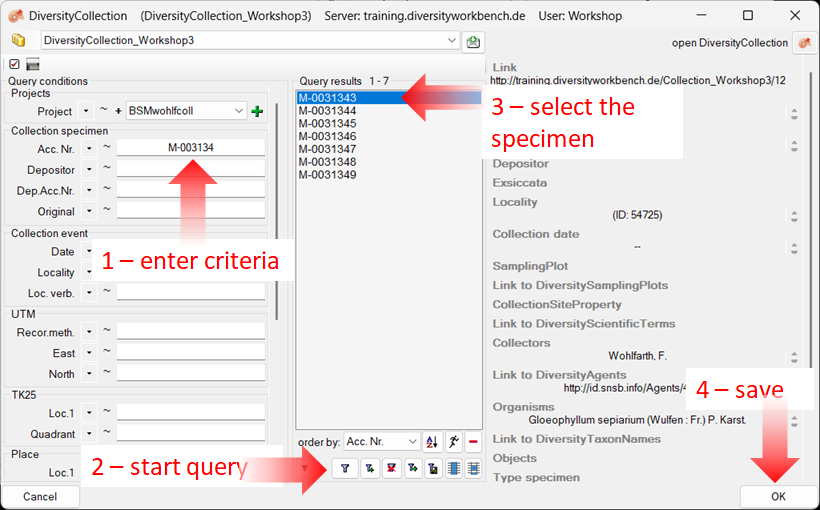
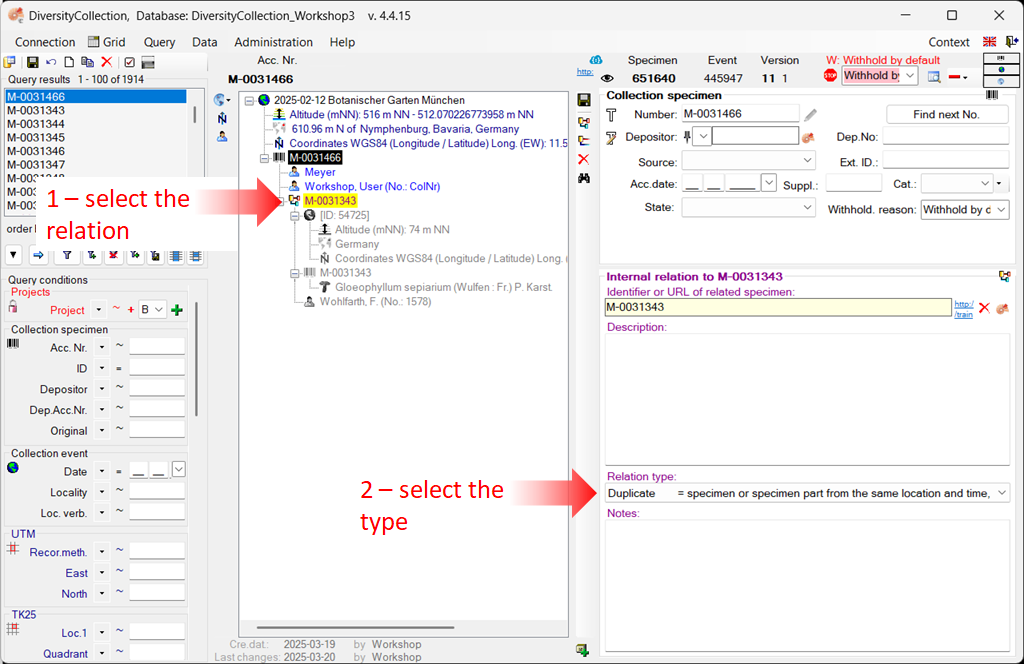
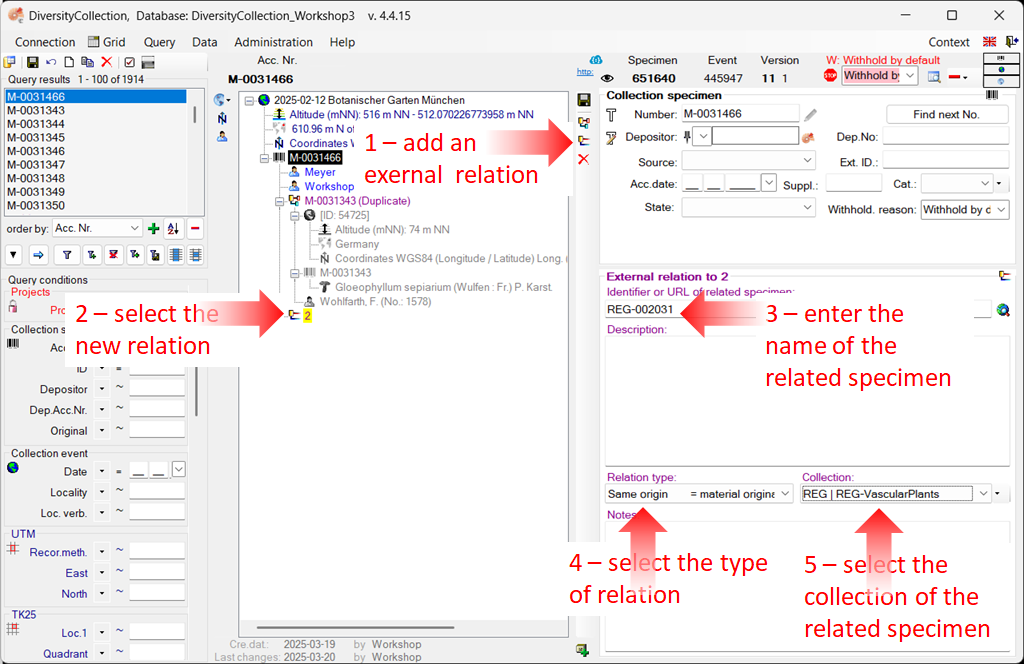
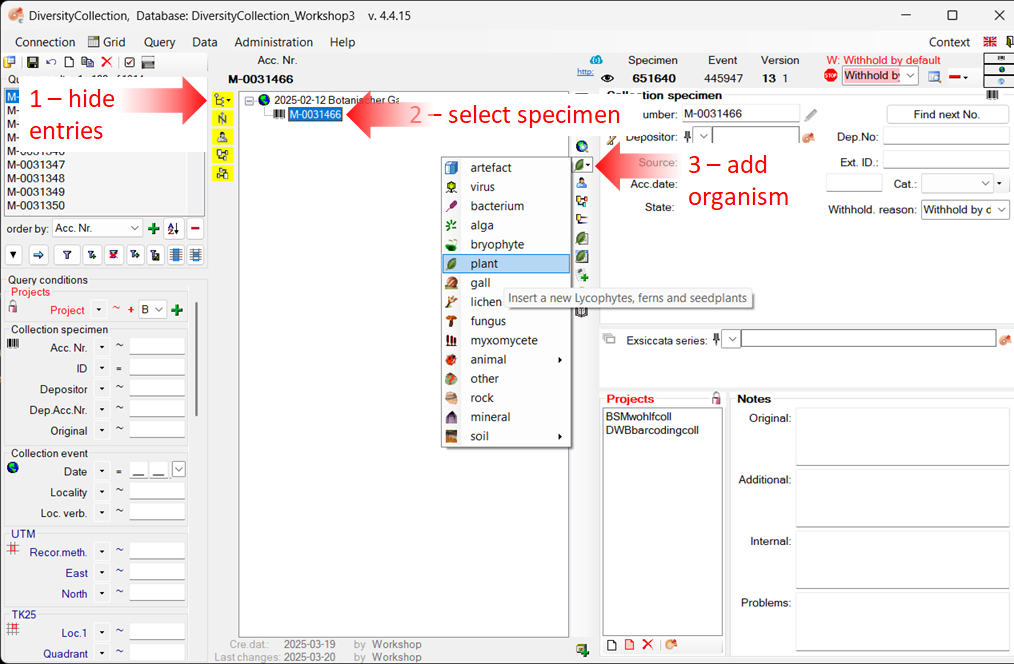
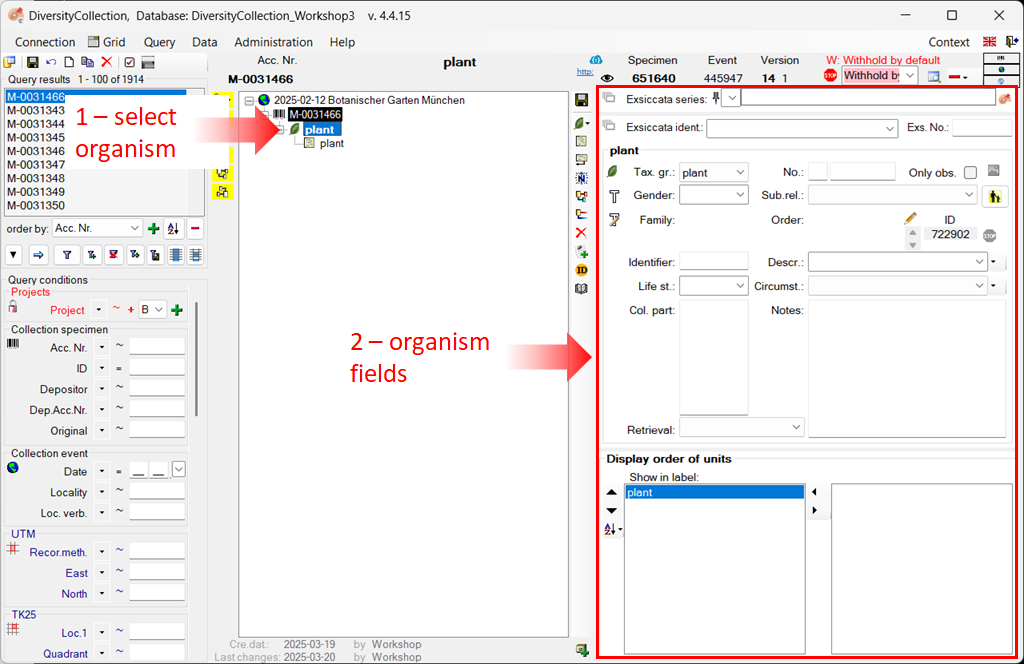
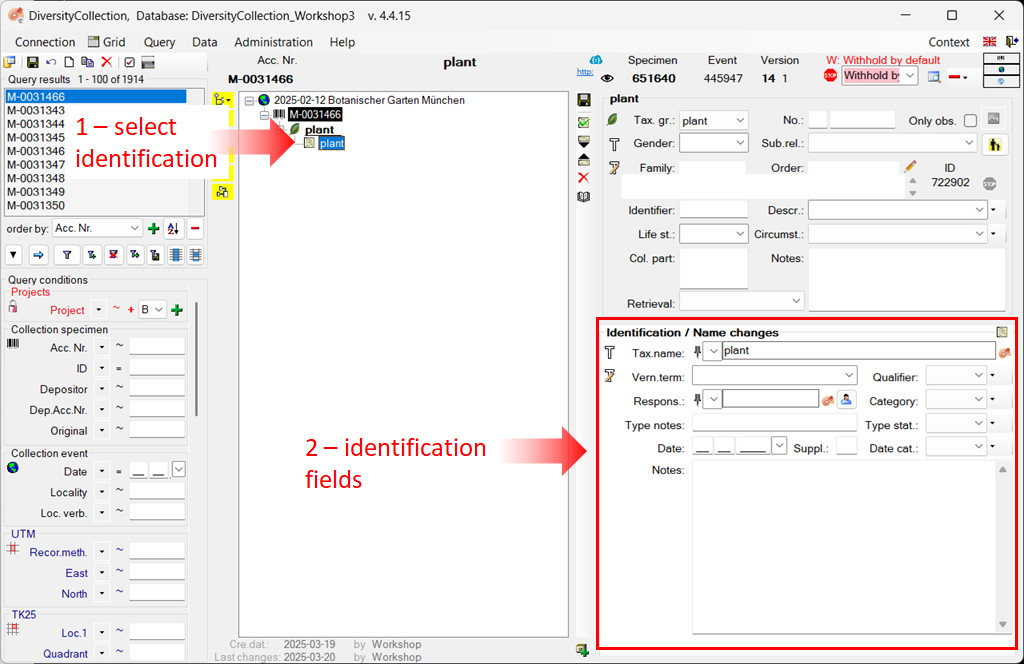
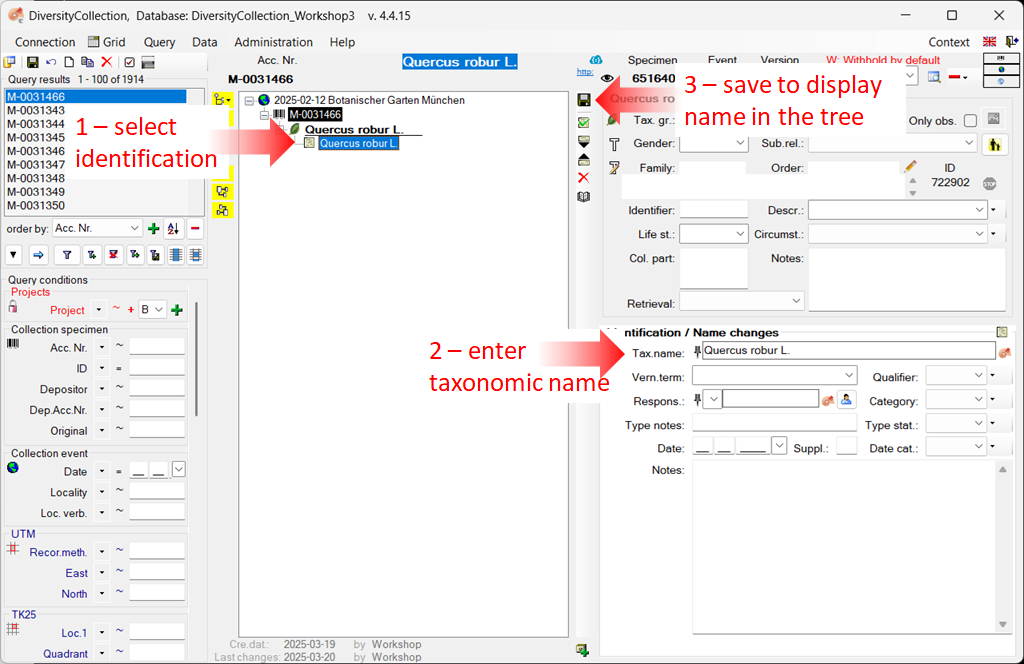
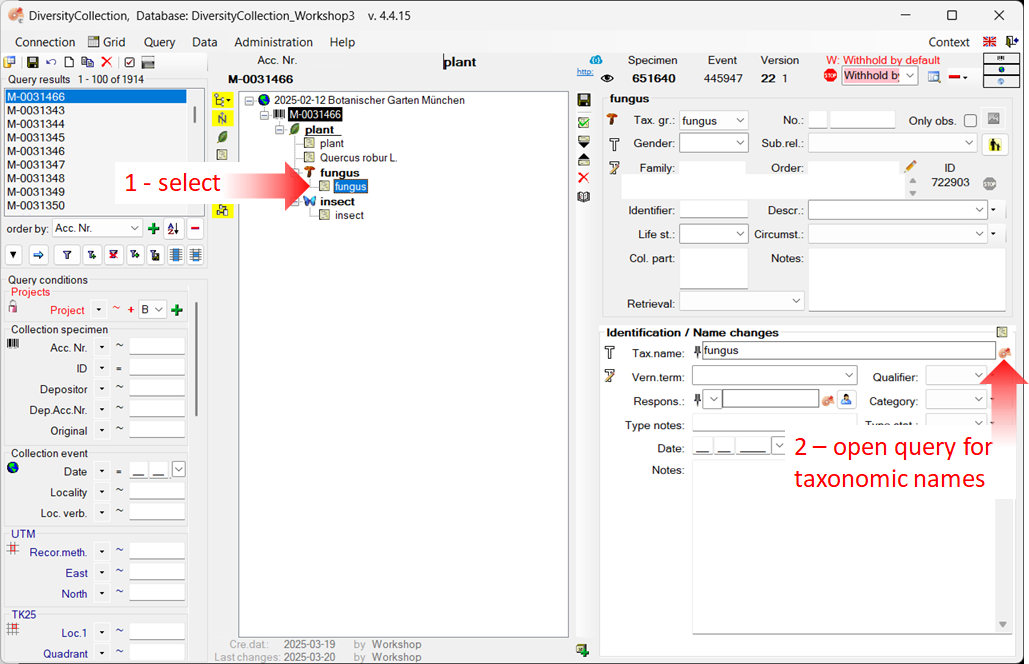
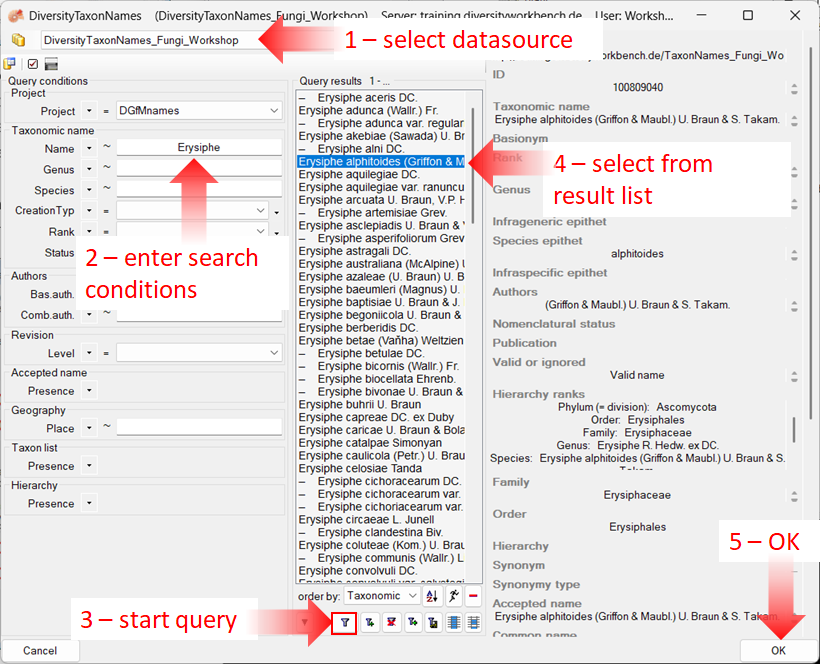
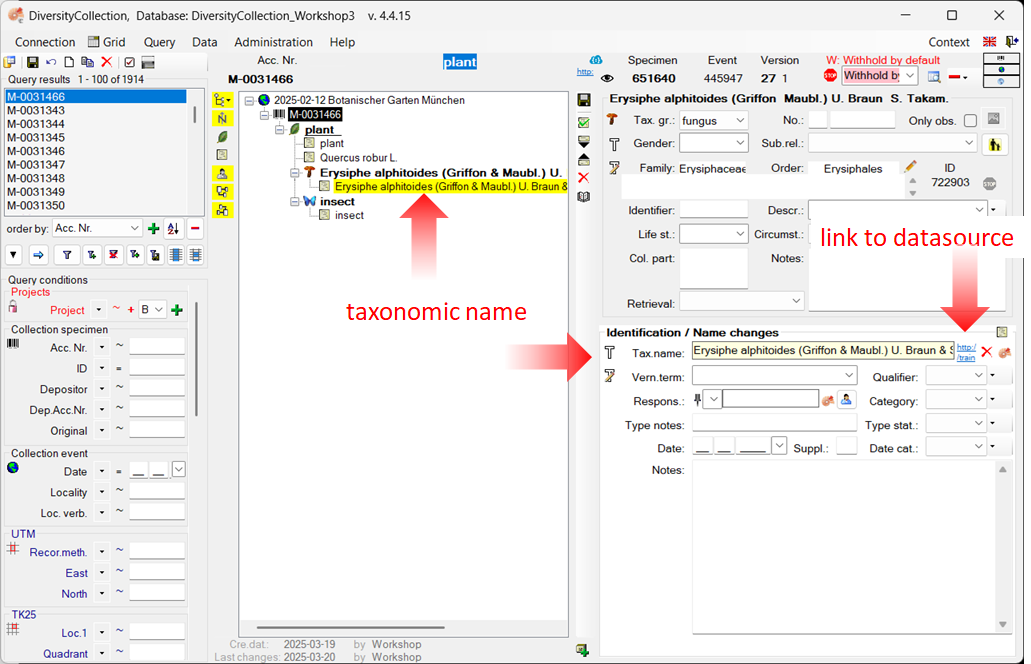
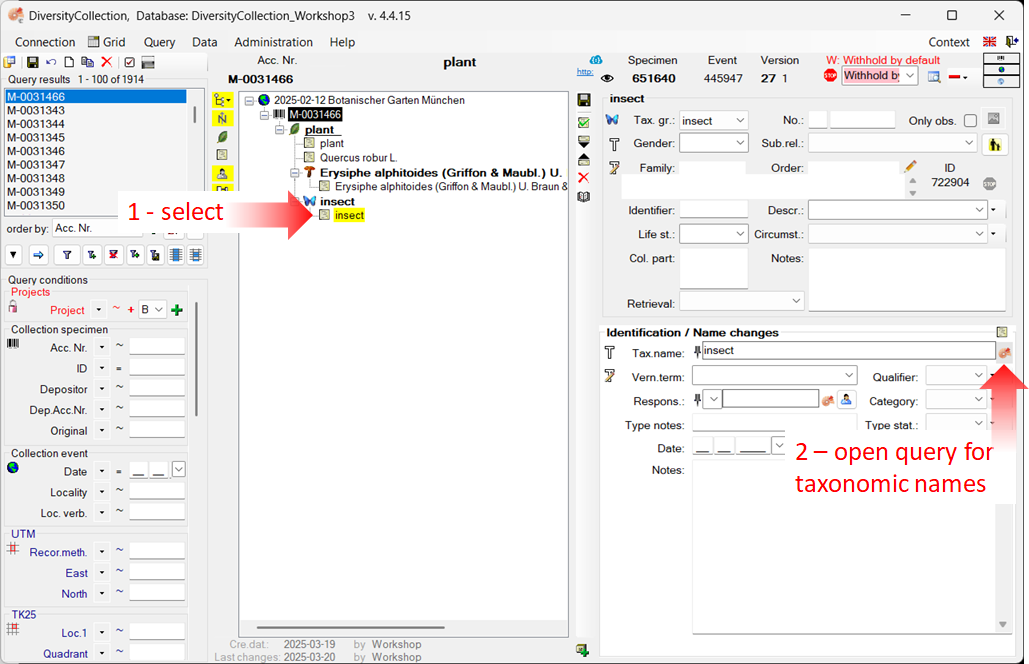
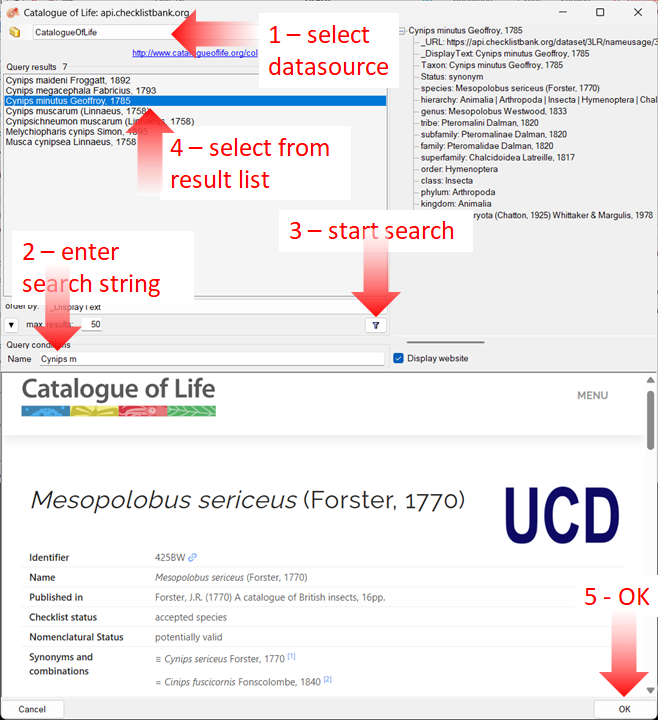
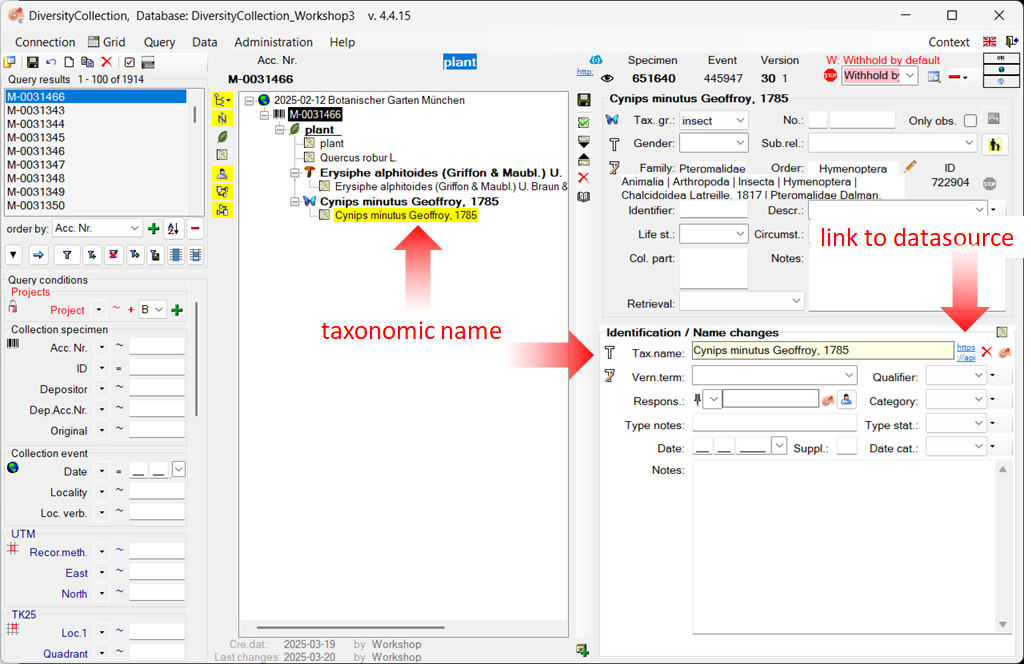
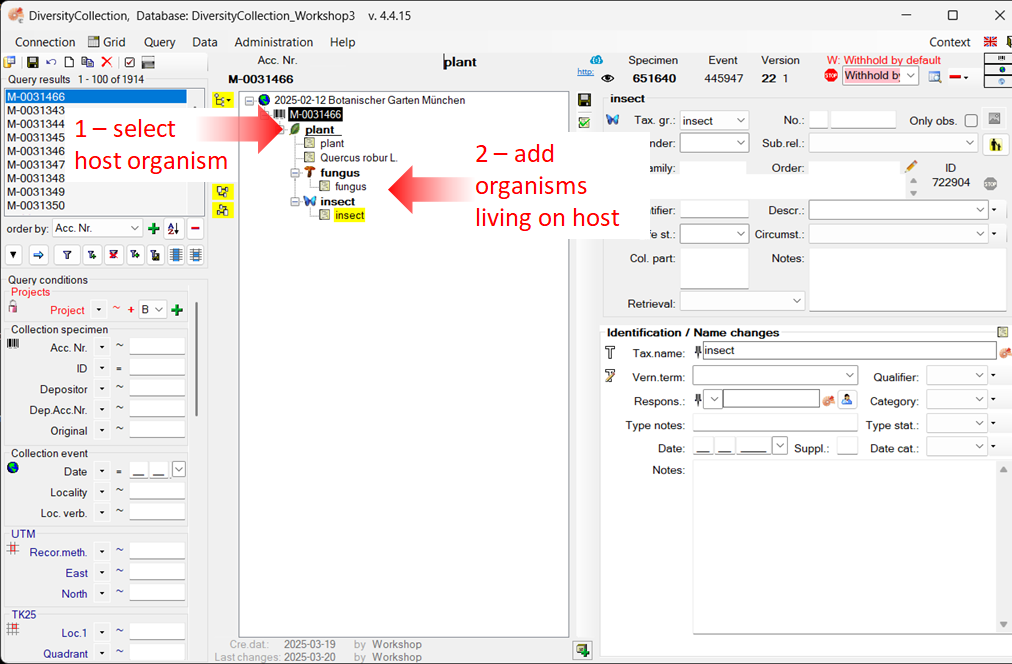
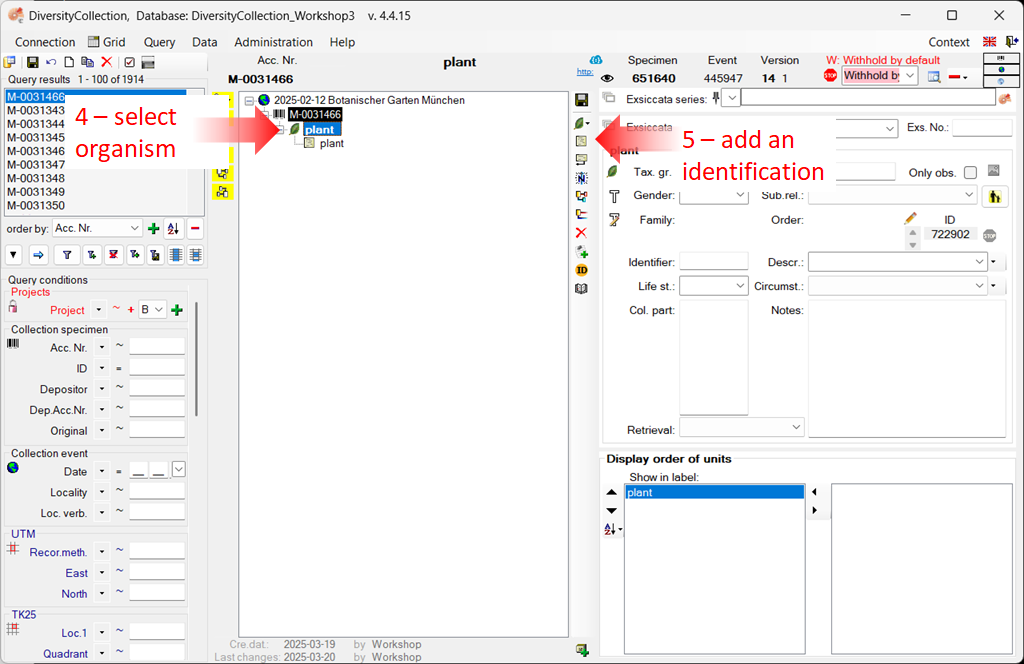
 located beneath the specimen tree (see point 1 in the image below).
located beneath the specimen tree (see point 1 in the image below).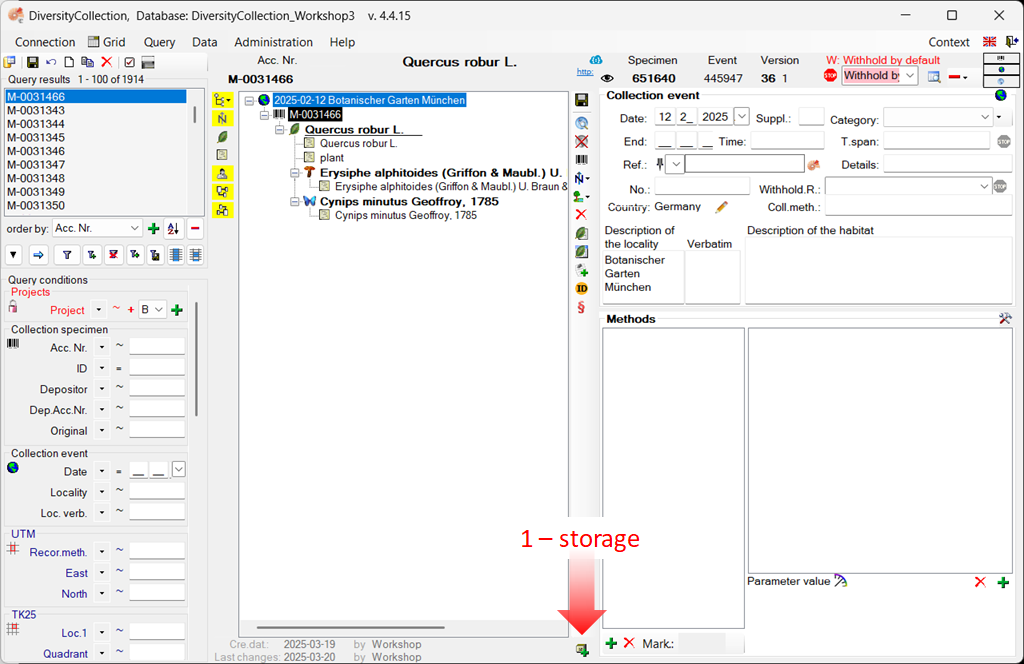

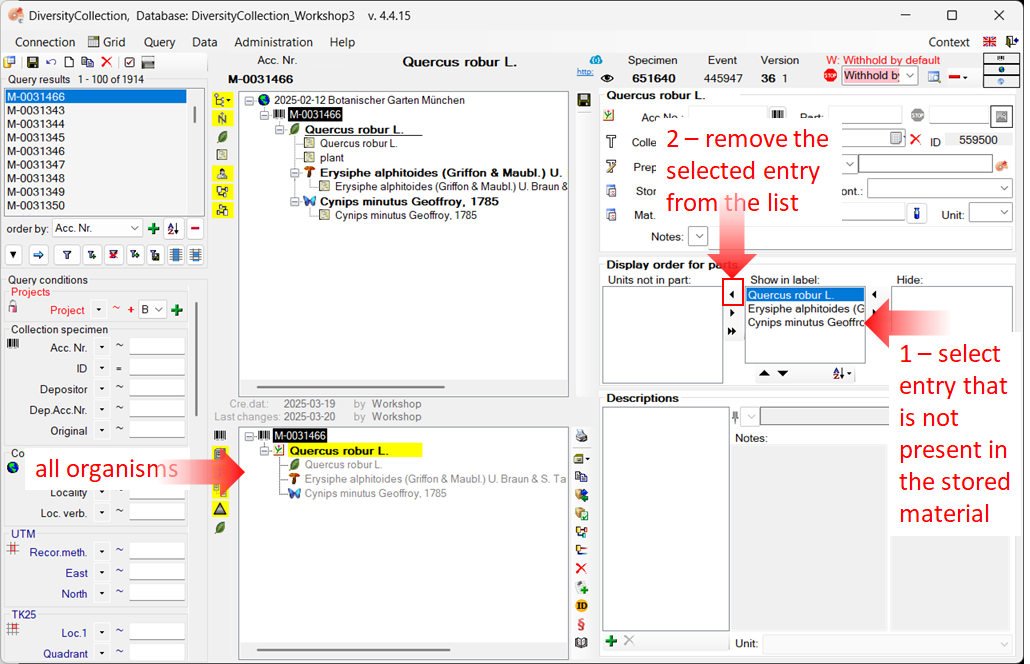
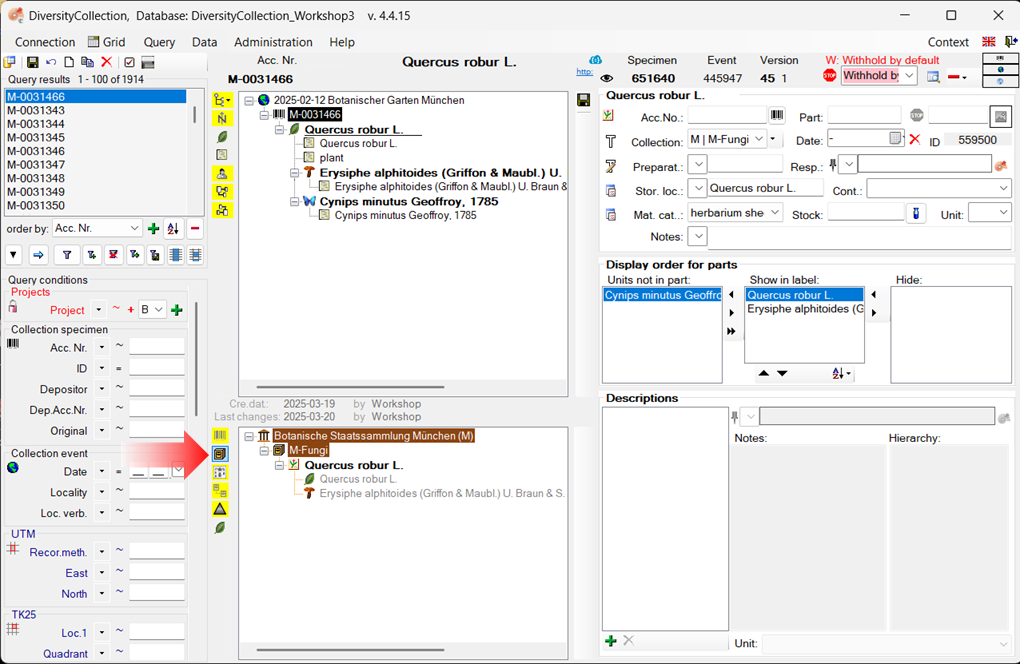
 Printing (see point 2 in the image below).
Printing (see point 2 in the image below).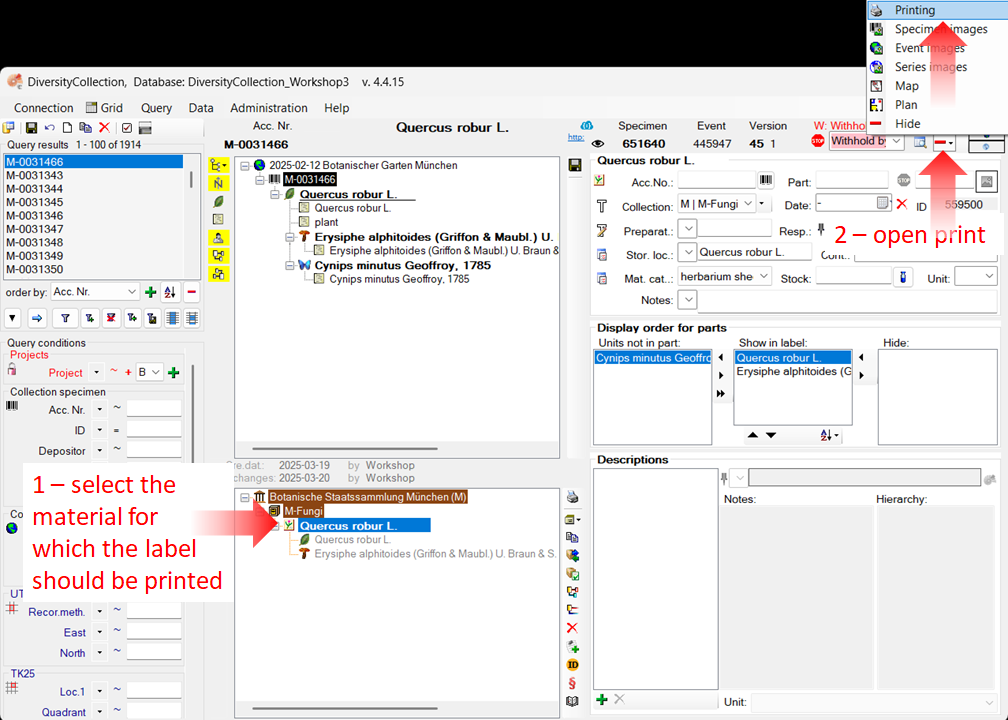
 (see point 3) to select a schema file (e.g. Standard.xslt).
(see point 3) to select a schema file (e.g. Standard.xslt).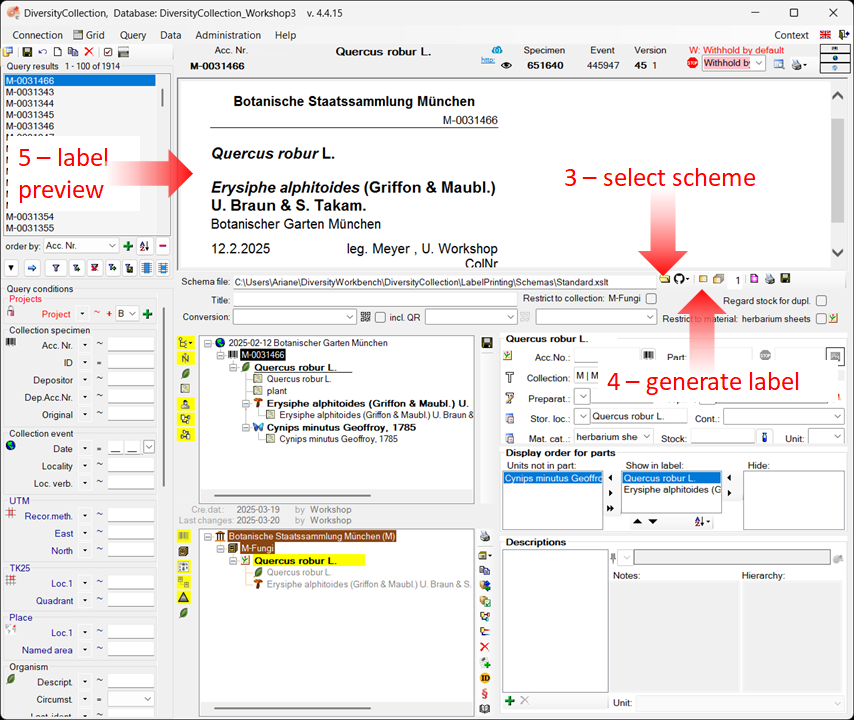
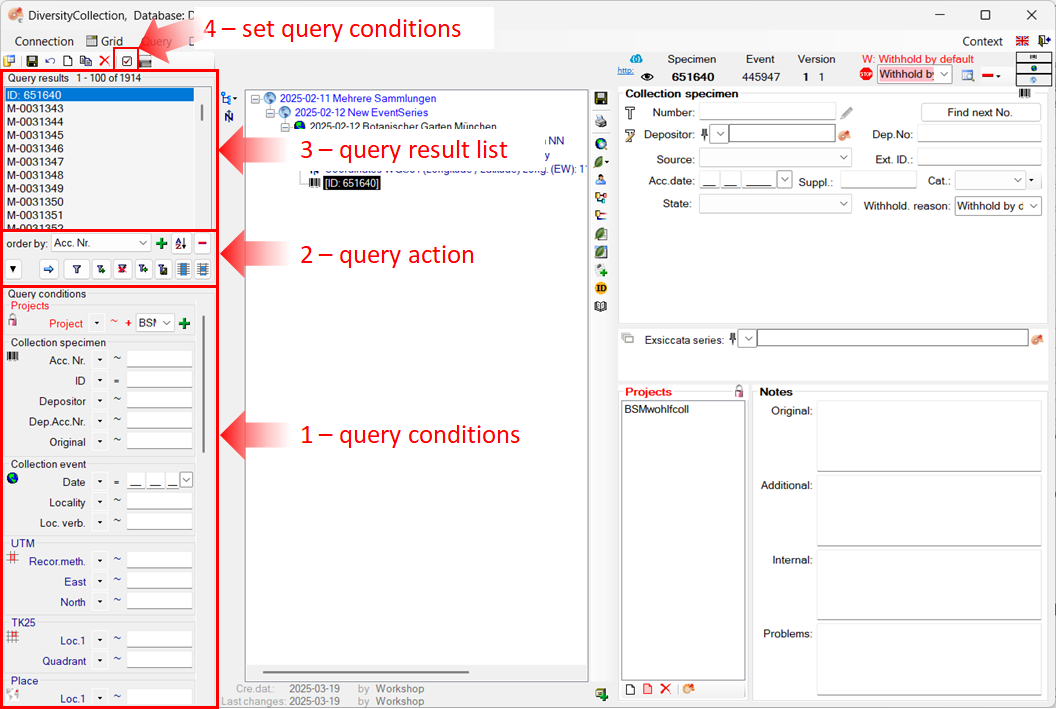
 in the top panel (see point 4 in the image above).
in the top panel (see point 4 in the image above).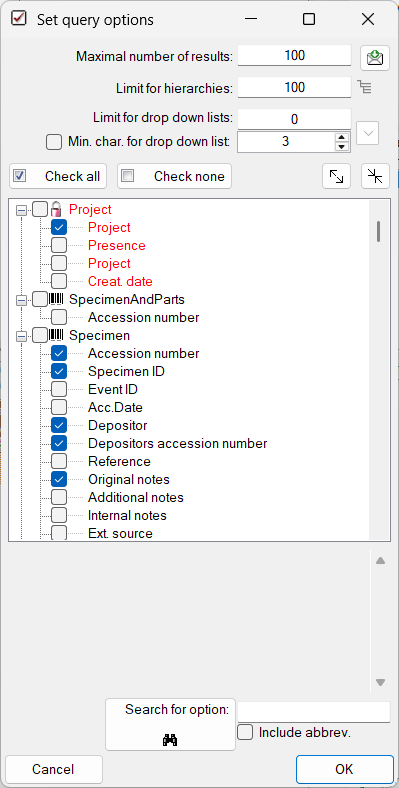
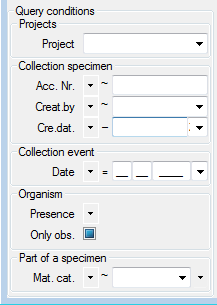
 button on the right side of the combobox to open the hierarchy and select
“herbarium sheet”.
button on the right side of the combobox to open the hierarchy and select
“herbarium sheet”. remember query option is active, the last query will be saved and is available after restarting the application.
remember query option is active, the last query will be saved and is available after restarting the application.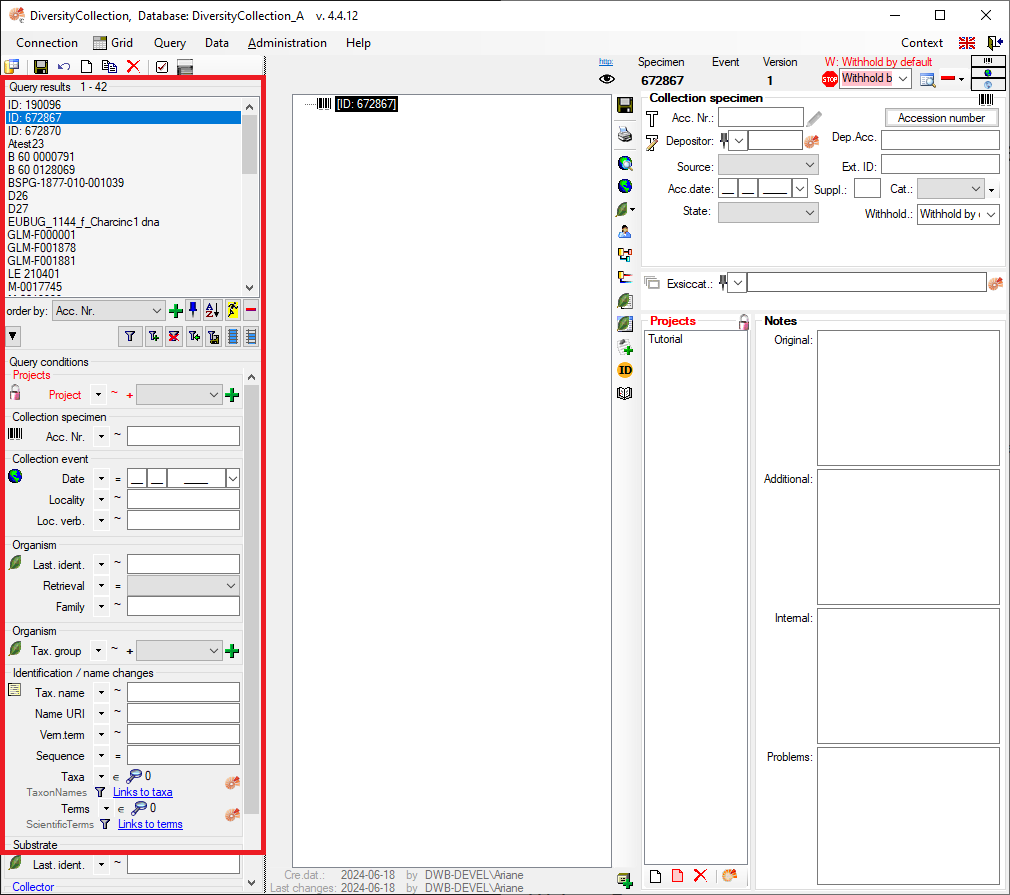
 .
.
 .
. .
. , Reference
, Reference
 . Remove a ‘duplicate’ search condition with a click on the red “Minus” button
. Remove a ‘duplicate’ search condition with a click on the red “Minus” button 



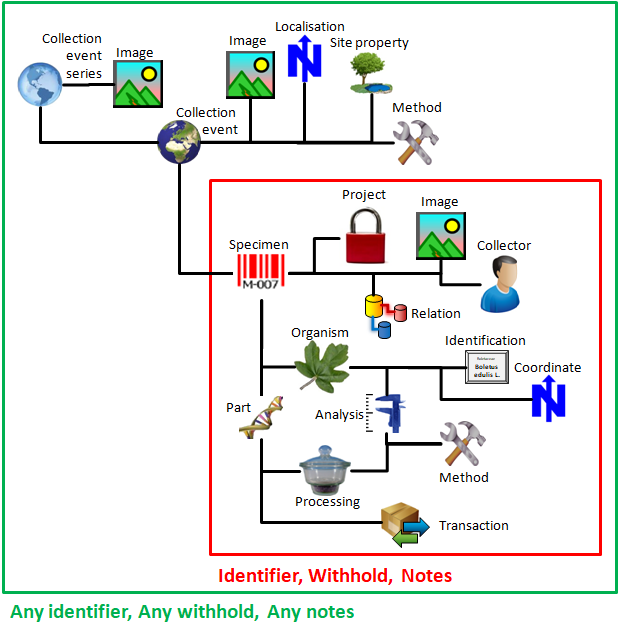
 Optimized query:
Optimized query: 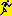
 . To display
several columns in the result list, click on the
. To display
several columns in the result list, click on the  button.
To change the sorting of the added order column click on the
button.
To change the sorting of the added order column click on the
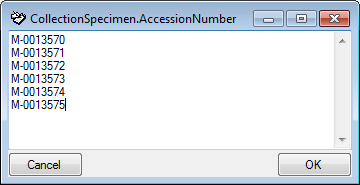
 button.
button.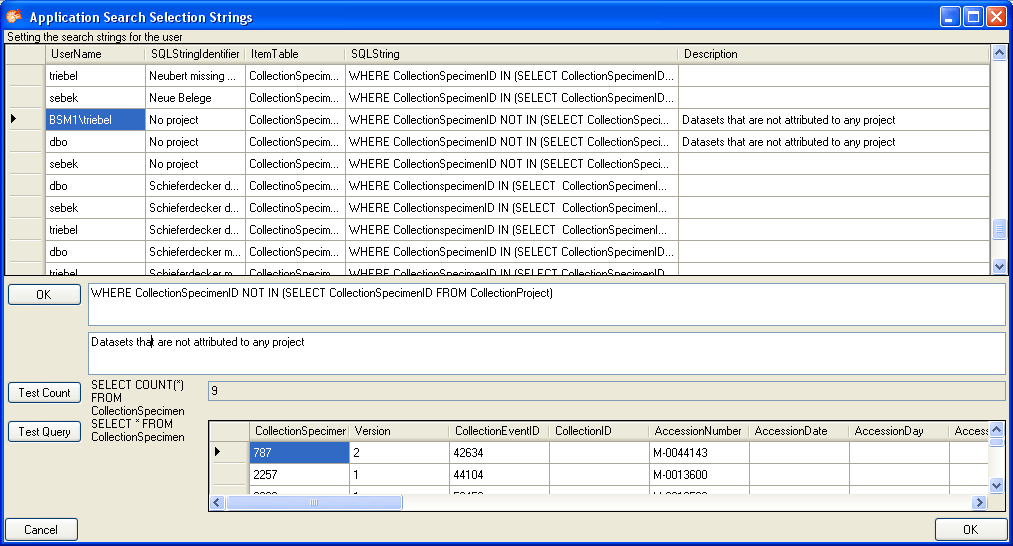
 = no,
= no,
 = undefined
= undefined button. A window will open as shown below where you may specify the
title and description of you query.
button. A window will open as shown below where you may specify the
title and description of you query.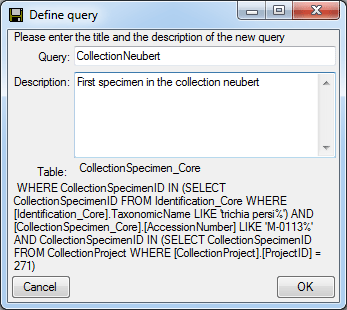
 icon and yellow background (see below).
icon and yellow background (see below).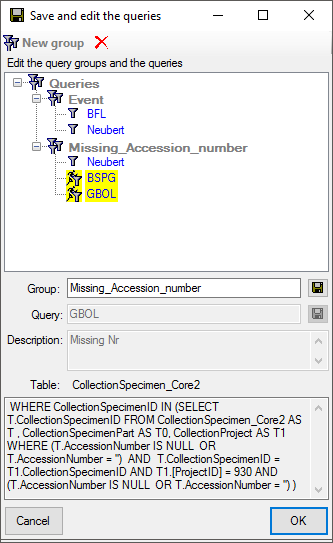
 button. A window will open as shown
below. Please note that if the
button. A window will open as shown
below. Please note that if the 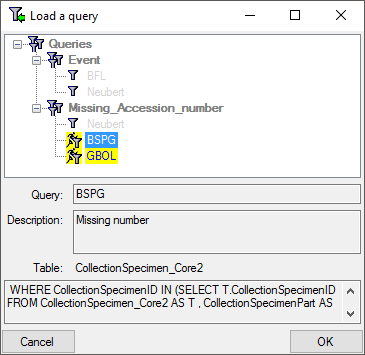
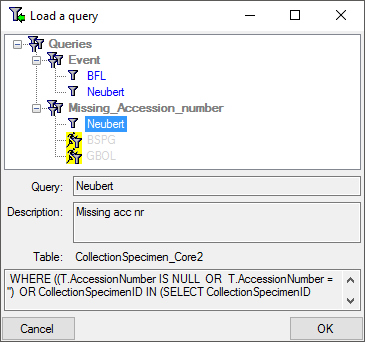
 or the
identifier of the organism
or the
identifier of the organism  . If the field
for the entry of the identifier
. If the field
for the entry of the identifier 
 button above the
button above the 

 button. If you wish to remove entries from
the selected list, choose them in the list and click on the
button. If you wish to remove entries from
the selected list, choose them in the list and click on the
 .
.

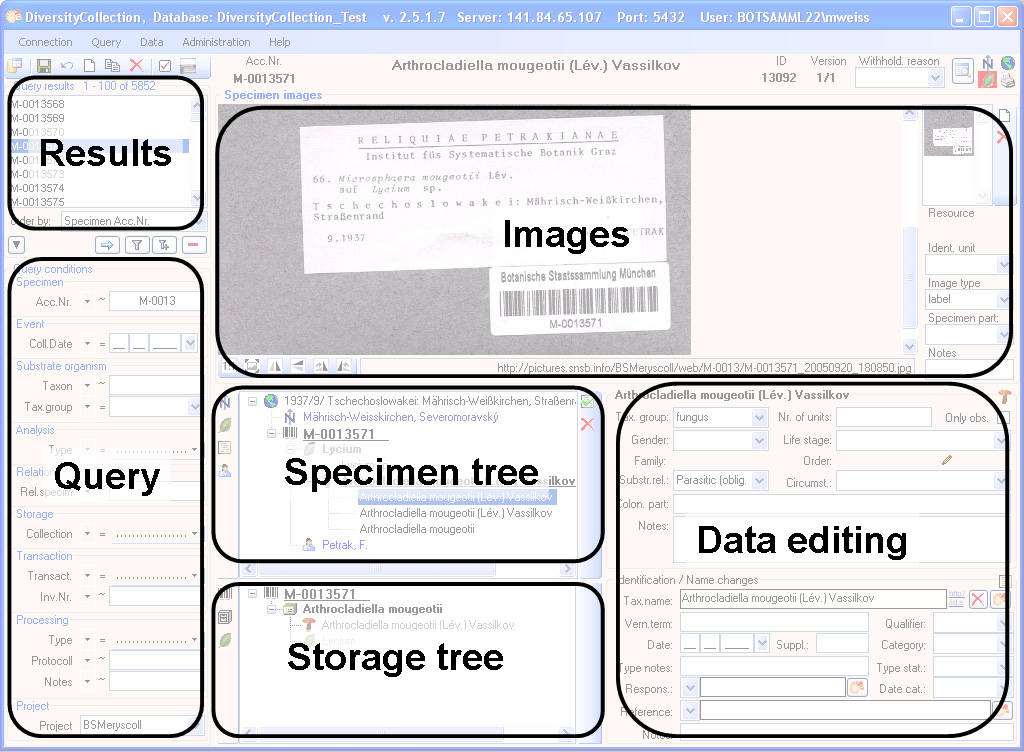
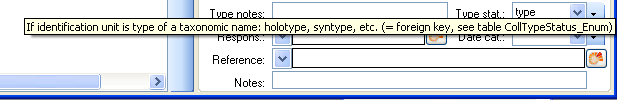


 the
the 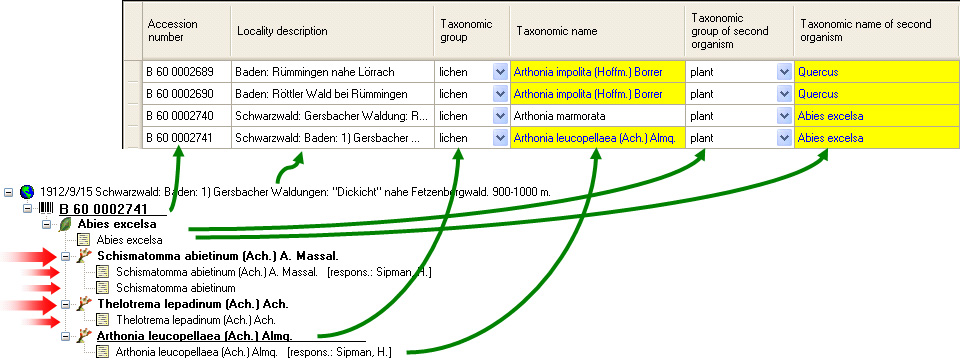
 button for the optimal height of the rows or the
button for the optimal height of the rows or the  button for the optimal width of the columns. To change the
width and sequence of the columns, drag the
columns to the position of your choice or adjust the width to your
preference. These changes will be saved for this session and
the next time you use grid mode. To return to the original sequence
of the columns, click the Reset sequence button.
button for the optimal width of the columns. To change the
width and sequence of the columns, drag the
columns to the position of your choice or adjust the width to your
preference. These changes will be saved for this session and
the next time you use grid mode. To return to the original sequence
of the columns, click the Reset sequence button.
 button to select the whole column. You can then select the function you
wish to use (remove, insert, append or replace). To replace a part of
the text in the selected fields, type the text to be replaced
and type the replacement in the appropriate fields. To start the
replacement, click on the
button to select the whole column. You can then select the function you
wish to use (remove, insert, append or replace). To replace a part of
the text in the selected fields, type the text to be replaced
and type the replacement in the appropriate fields. To start the
replacement, click on the  button. To append a string to all entries
in the selected fields, click the
button. To append a string to all entries
in the selected fields, click the  button. To
remove all entries from the selected fields, click the
button. To
remove all entries from the selected fields, click the  button.
button.

 . Simply
click on the button associated with a link to remove the connection to the
corresponding module. After doing so, you can edit the text field containing
the previously linked value.
. Simply
click on the button associated with a link to remove the connection to the
corresponding module. After doing so, you can edit the text field containing
the previously linked value. button. To undo
all changes since the last time the datasets were saved, click the
button. To undo
all changes since the last time the datasets were saved, click the  button. To save the changes to the current dataset
use the
button. To save the changes to the current dataset
use the  undo button. If you click
the OK button, you will be asked if you want to save the changes before
the window closes. If you click the Cancel button or close the window, your changes will not be
saved. To export the data displayed in the grid as a text file with tabs as
column separators, click on the
undo button. If you click
the OK button, you will be asked if you want to save the changes before
the window closes. If you click the Cancel button or close the window, your changes will not be
saved. To export the data displayed in the grid as a text file with tabs as
column separators, click on the 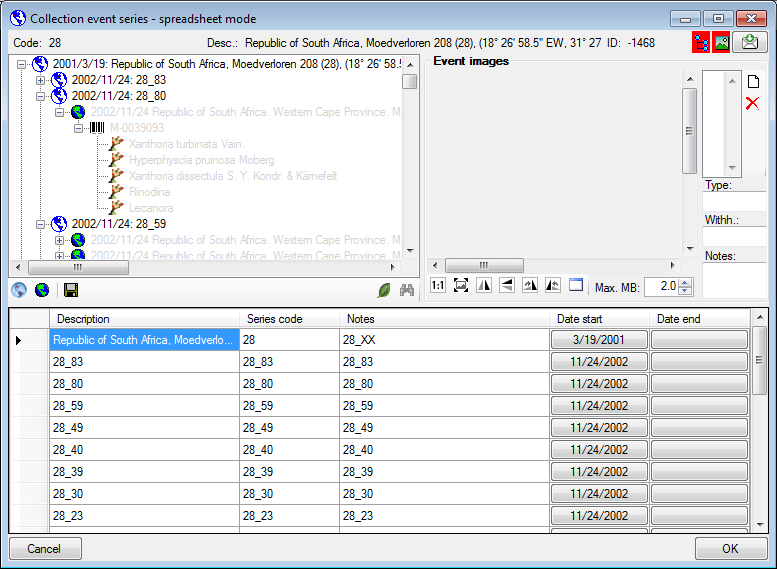
 button for the hierarchy tree and
the
button for the hierarchy tree and
the  .
. button. A window will open as described in the
button. A window will open as described in the
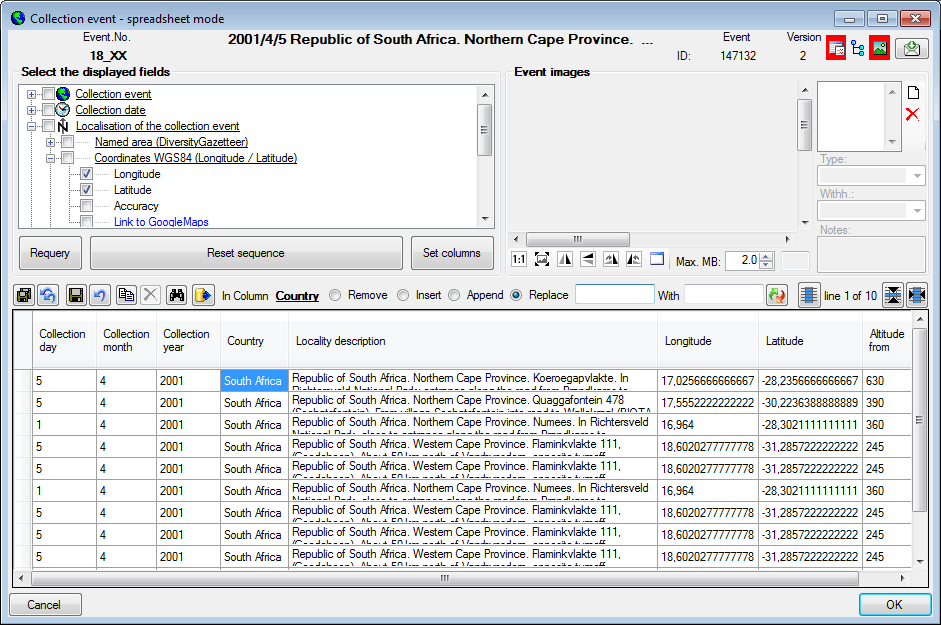
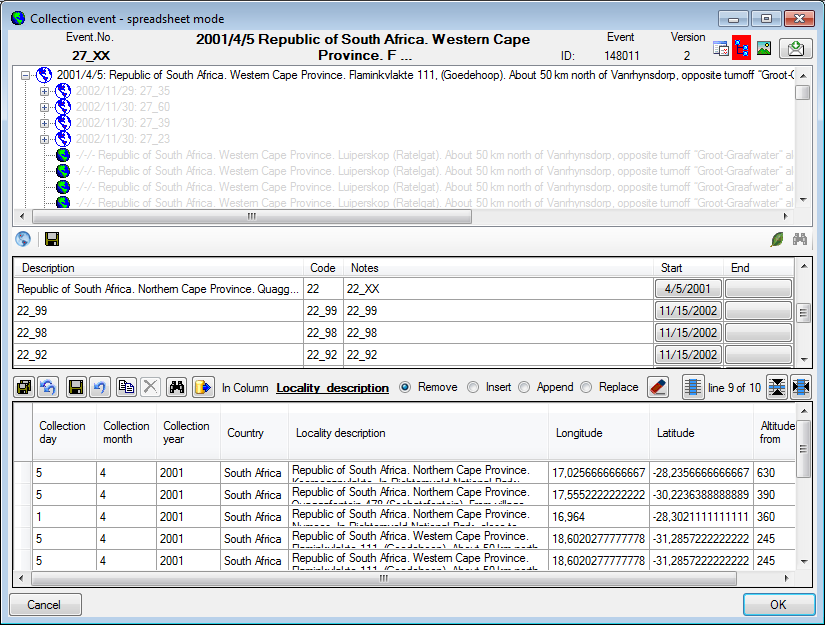
 (the column
[Related specimen URL] will be shown in the interface)
or internal.
(the column
[Related specimen URL] will be shown in the interface)
or internal.  (the column
[Related specimen display text] will be shown in the interface)
Use the [Relation is internal] column to change the state and
the [Link to DiversityCollection for relation] column to set an
internal relation.
(the column
[Related specimen display text] will be shown in the interface)
Use the [Relation is internal] column to change the state and
the [Link to DiversityCollection for relation] column to set an
internal relation. 

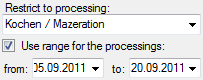
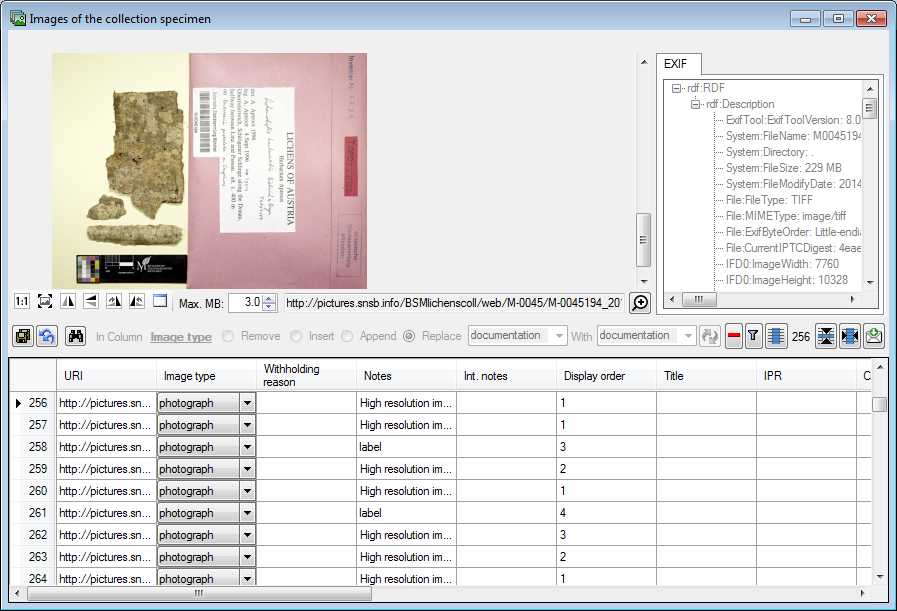
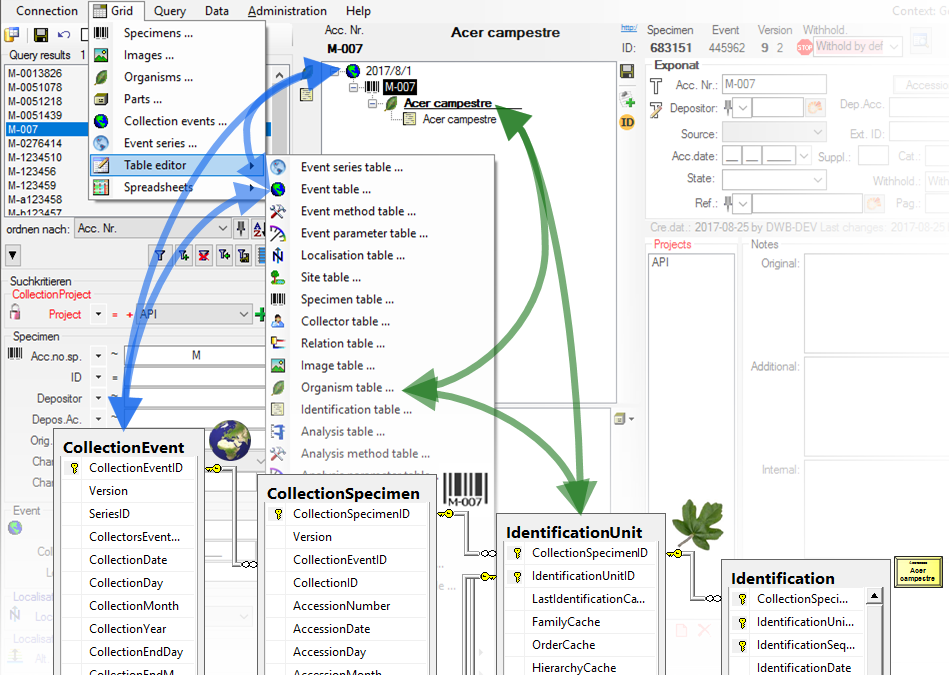
 button the width is set to the
content. After the column width is set, this will be indicated with a
yellow background
button the width is set to the
content. After the column width is set, this will be indicated with a
yellow background  . Click again on the
. Click again on the 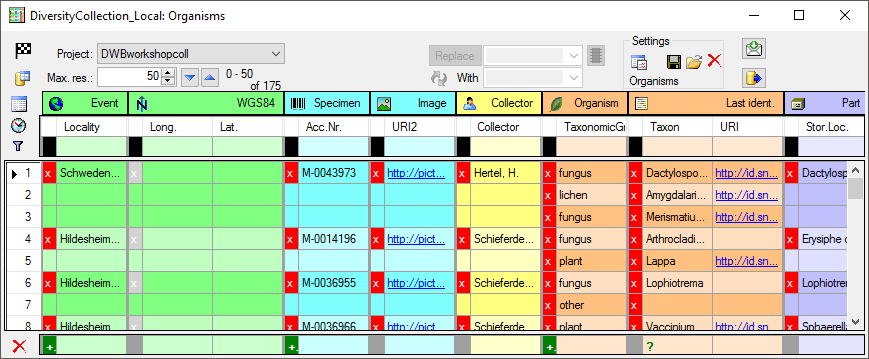
 button. Now the application will directly open the spreadsheet with the same parameters when you close it.
button. Now the application will directly open the spreadsheet with the same parameters when you close it.  button will appear in front of the project. Click on this button to change to the list of projects with read only access. The window will change into the Read only mode (see below). To return to the list of projects with write access, just click no the button again. After changing the source for the project list, the project label will blink with red to remind you, to select a project from the list.
button will appear in front of the project. Click on this button to change to the list of projects with read only access. The window will change into the Read only mode (see below). To return to the list of projects with write access, just click no the button again. After changing the source for the project list, the project label will blink with red to remind you, to select a project from the list.  button.
button. 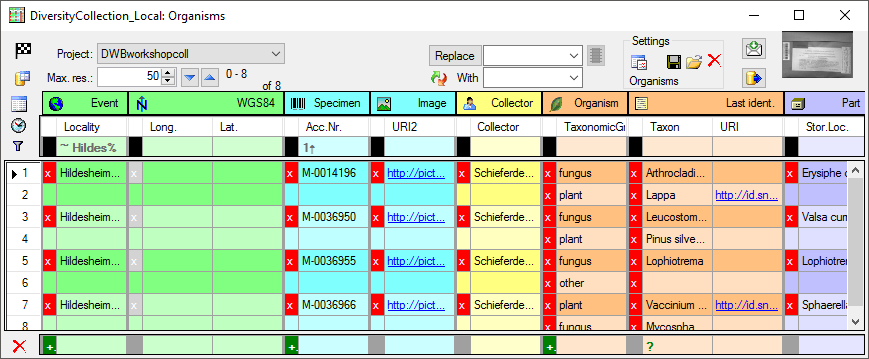
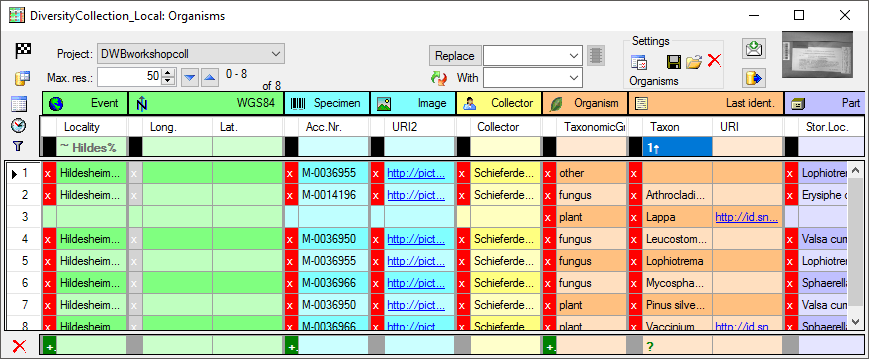
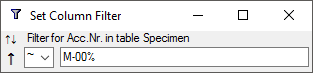
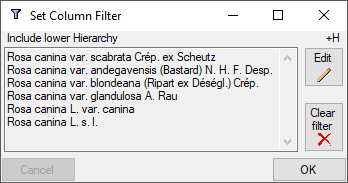
 button. Once you have selected the
contents to change, select the mode of change:
button. Once you have selected the
contents to change, select the mode of change: Overwrite: Remove current content and insert given value
Overwrite: Remove current content and insert given value Tranfer or
Tranfer or  Insert the content
of the clipboard into the selected cells.
Insert the content
of the clipboard into the selected cells. 
 and if the column has no relation to a module, the button will appear like
and if the column has no relation to a module, the button will appear like 
 checkbox
checkbox  checkbox. Displayed columns are indicated by a yellow background while hidden columns get a light yellow background. Certain columns are required (indicated by a purple color) and can not be removed e.g. if they are involved in a link to a remote module like in the example below the columns FamilyCache and OrderCache. Columns linked to a module are indicated by a blue color.
checkbox. Displayed columns are indicated by a yellow background while hidden columns get a light yellow background. Certain columns are required (indicated by a purple color) and can not be removed e.g. if they are involved in a link to a remote module like in the example below the columns FamilyCache and OrderCache. Columns linked to a module are indicated by a blue color.
 button allows you to include one of the tables missing in the sheet (see image below). After selection of the table, select the column(s) you want to include in the sheet as shown in the image above.
button allows you to include one of the tables missing in the sheet (see image below). After selection of the table, select the column(s) you want to include in the sheet as shown in the image above.
 Resources directory ... . For an introduction see a short tutorial
Resources directory ... . For an introduction see a short tutorial
 button. A window as shown below will open, where
you can set the parameters for the map.
button. A window as shown below will open, where
you can set the parameters for the map.  Symbols:
Symbols: 
 button
to select the source table. The programm will try to find the relevant
data an make a proposal as shown in the image below.
button
to select the source table. The programm will try to find the relevant
data an make a proposal as shown in the image below.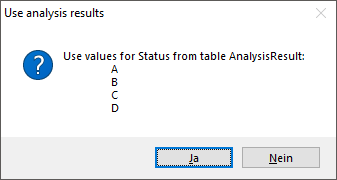
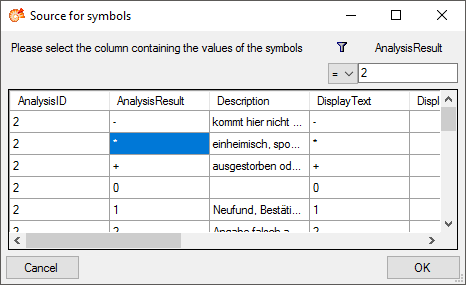
 and will
now remove the source, to return to the values contained in the
spreadsheet. If certain values should not appear in the map, choose
and will
now remove the source, to return to the values contained in the
spreadsheet. If certain values should not appear in the map, choose
 legend:
legend: 
 Gazetteer for
retrieval of TK25 coordinates. The coordinates for the symbols
will be obtained from a gazetteer module. Please select the source as
shown in the example below.
Gazetteer for
retrieval of TK25 coordinates. The coordinates for the symbols
will be obtained from a gazetteer module. Please select the source as
shown in the example below.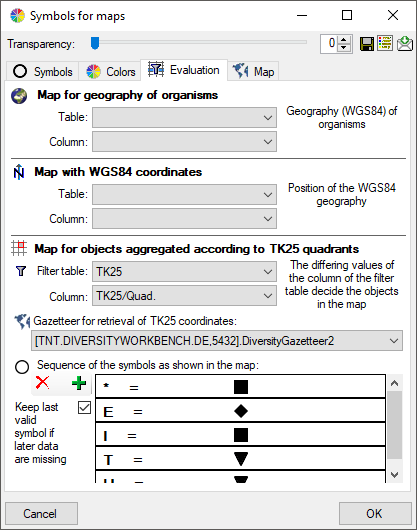
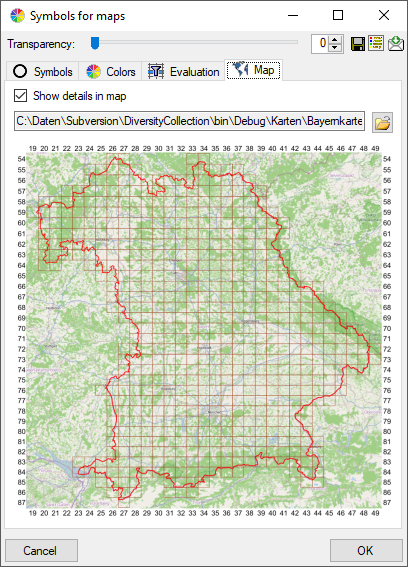
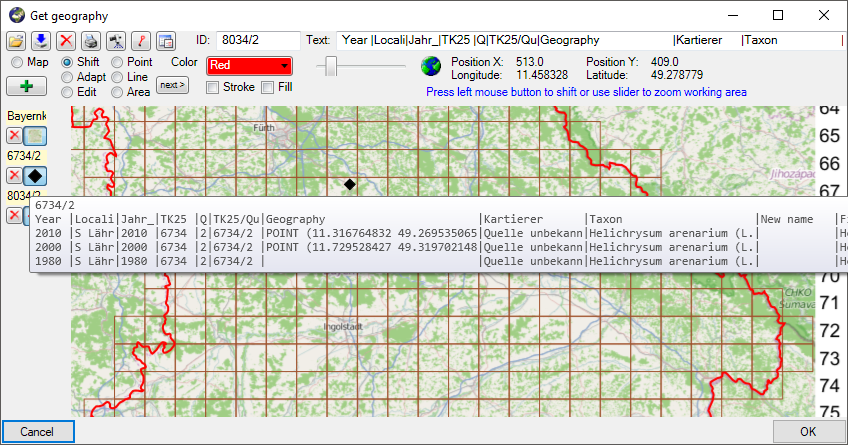
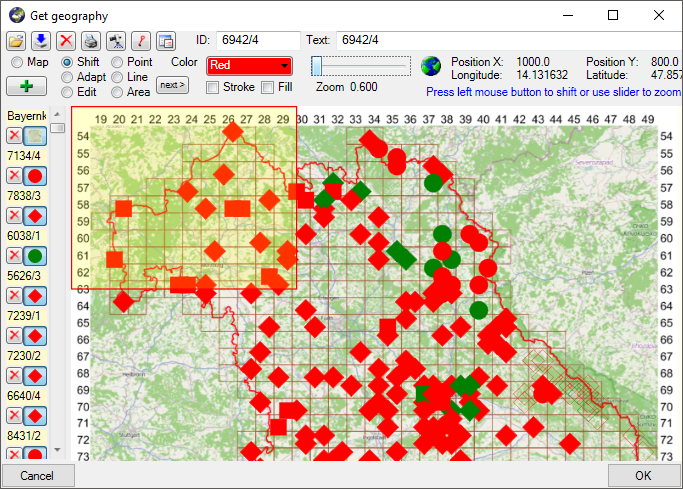
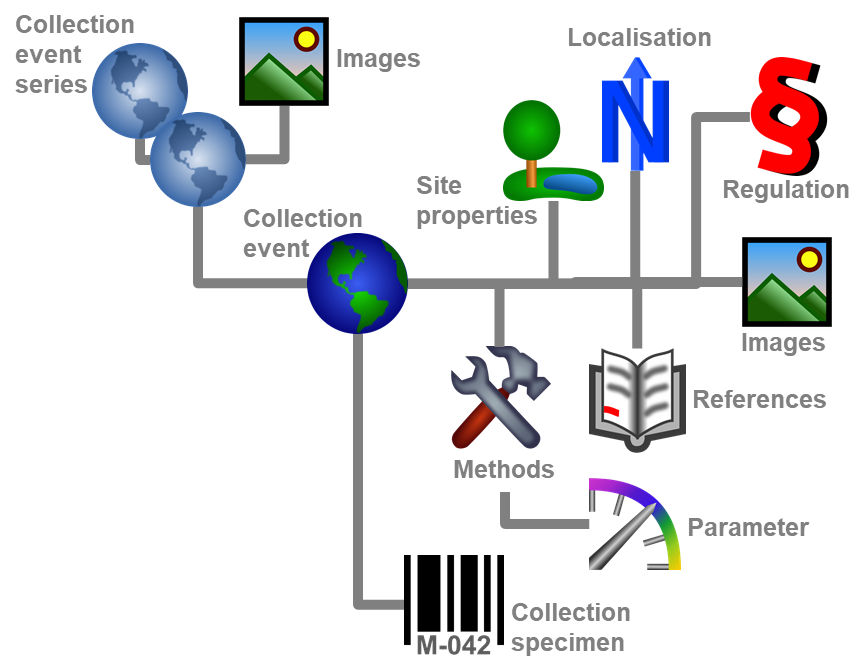


 button.
button.
 you can switch to a new Chromium-based browser for the display of
images. The presence of images in an event series is indicated by a
yellow background on the lower of the buttons in the upper right
corner. These buttons may be used as a shortcut to display the
images
you can switch to a new Chromium-based browser for the display of
images. The presence of images in an event series is indicated by a
yellow background on the lower of the buttons in the upper right
corner. These buttons may be used as a shortcut to display the
images  .
.
 button. To adapt the size of the image to the available space in the
form, click on the
button. To adapt the size of the image to the available space in the
form, click on the  flip horizontally,
flip horizontally,
 flip vertically,
flip vertically,
 rotate right,
rotate right,
 rotate left). To view the image in a
separate form, click on the
rotate left). To view the image in a
separate form, click on the  Map in the header to
show the maps and use the
Map in the header to
show the maps and use the  in the header menu to display the images of
the collection event. In case the event images are not displayed, a
yellow (image(s) present) resp. gray (no image) backgroud of the tiny
buttons at the upper right corner indicates the presence of images
in the header menu to display the images of
the collection event. In case the event images are not displayed, a
yellow (image(s) present) resp. gray (no image) backgroud of the tiny
buttons at the upper right corner indicates the presence of images
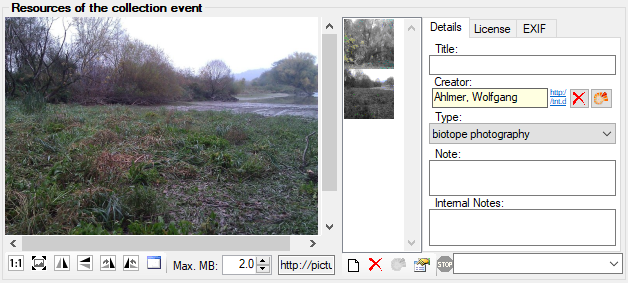


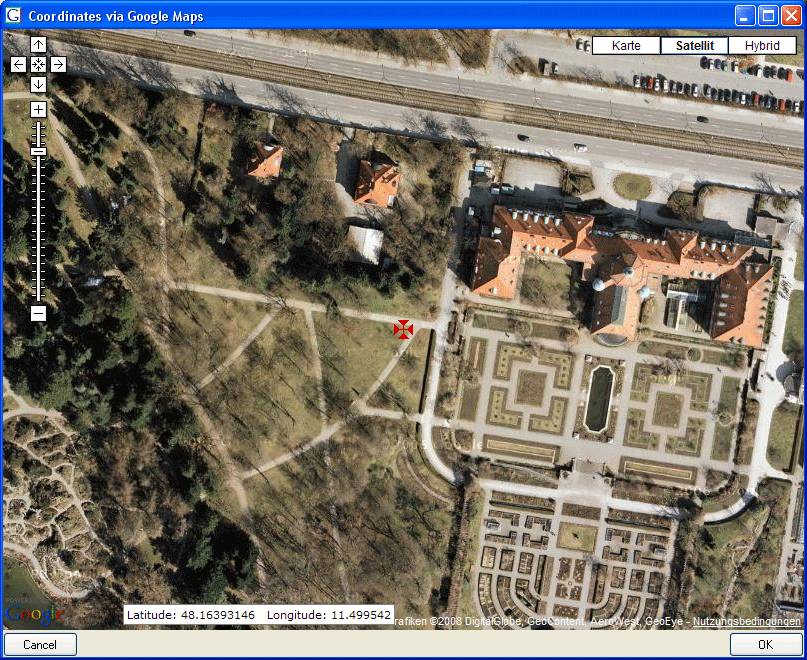


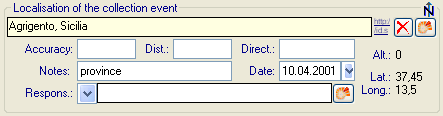

 button to incorporate the changed values into your data. The system will
calculate the corresponding value for storage in the database together
with the default accuracy. You may correct this accuracy if you have
more exact values. To indicate that the shown values are calculated from
the values in the database the fields have a green background. When you
save the data DiversityCollection will store geographic coordinates and
the average altitude, if available, in separate fields.
button to incorporate the changed values into your data. The system will
calculate the corresponding value for storage in the database together
with the default accuracy. You may correct this accuracy if you have
more exact values. To indicate that the shown values are calculated from
the values in the database the fields have a green background. When you
save the data DiversityCollection will store geographic coordinates and
the average altitude, if available, in separate fields. Available units: meter and feet.
Available units: meter and feet. Available units: Orientation (N, NE, ... ) and
degree rel. to North.
Available units: Orientation (N, NE, ... ) and
degree rel. to North. Available units: degree and percent.
Available units: degree and percent.
 GIS viewer or the
GIS viewer or the 
 . For the positions of the
organisms in this map, the ID of the organism ( = IdentificationUnitID,
528577 in example below) is shown as identifier and the last
identification ( = LastIdentificationCache) as display text (Ajuga
reptans L. in example below).
. For the positions of the
organisms in this map, the ID of the organism ( = IdentificationUnitID,
528577 in example below) is shown as identifier and the last
identification ( = LastIdentificationCache) as display text (Ajuga
reptans L. in example below).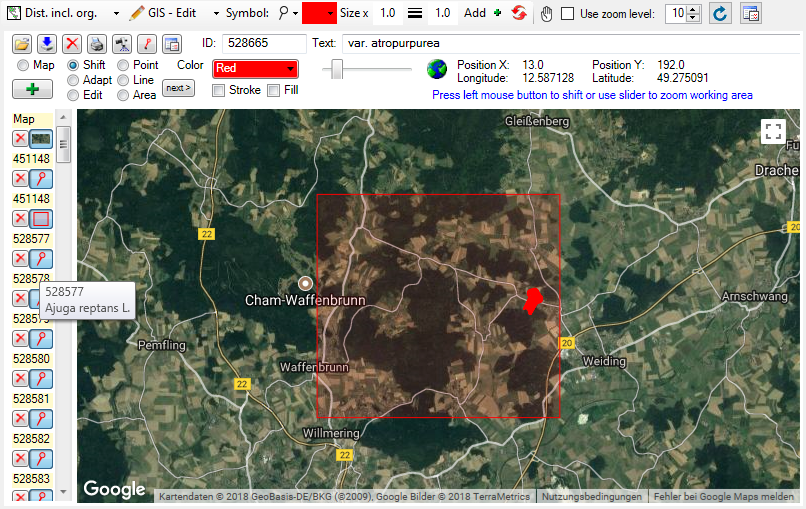


 button will show the entire hierarchy of
the SamplingPlots as shown below.
button will show the entire hierarchy of
the SamplingPlots as shown below.
 button will show the entire hierarchy of
the SamplingPlots as shown below.> button
you can retrieve a
button will show the entire hierarchy of
the SamplingPlots as shown below.> button
you can retrieve a 


 button. A window as shown below will open. To
select an item from the chart click no the entry you want to select and
close the window with a click on the OK button.
button. A window as shown below will open. To
select an item from the chart click no the entry you want to select and
close the window with a click on the OK button.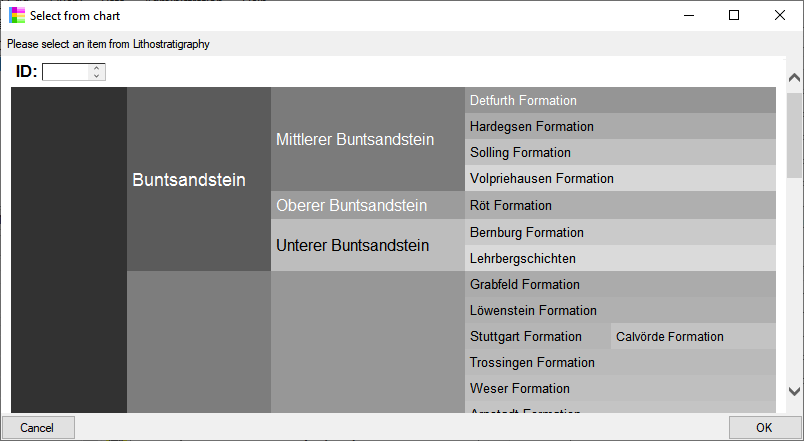
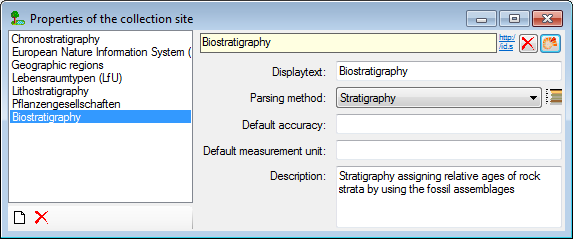
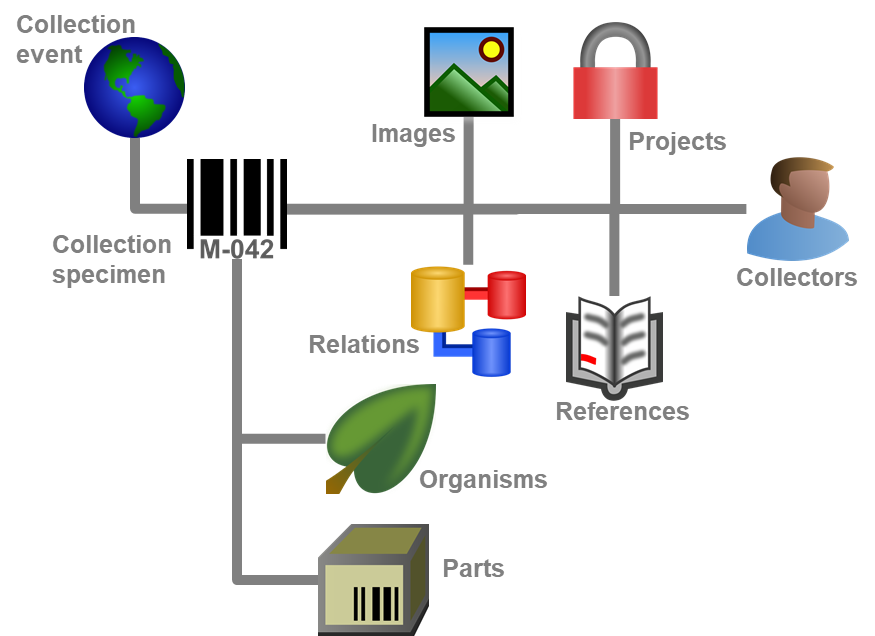


 images for the
specimen,
images for the
specimen, 
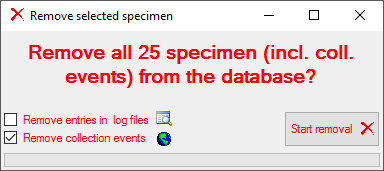
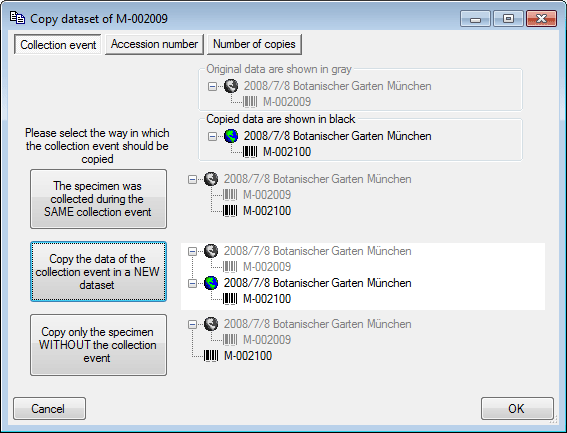
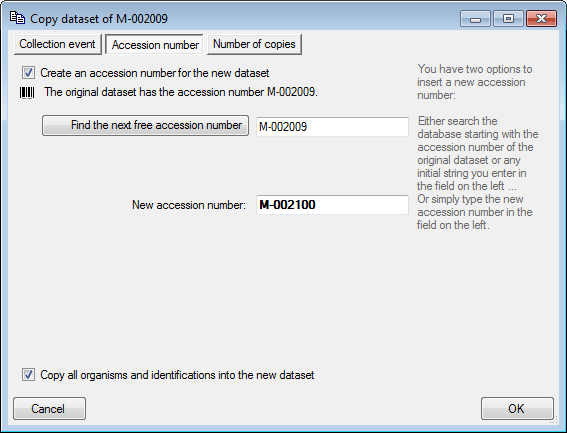
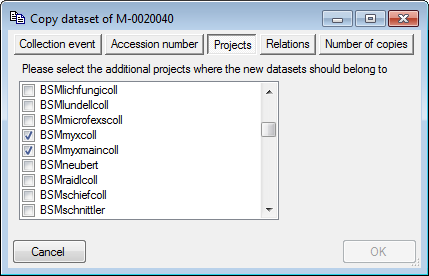
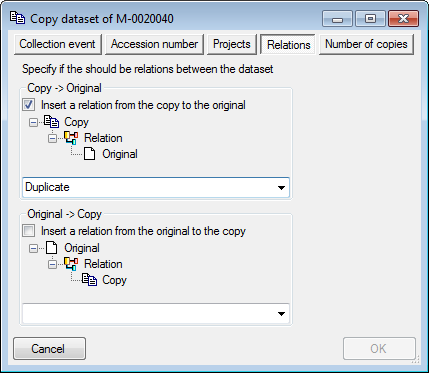
 None buttons to
select all resp. none of the tables.
None buttons to
select all resp. none of the tables.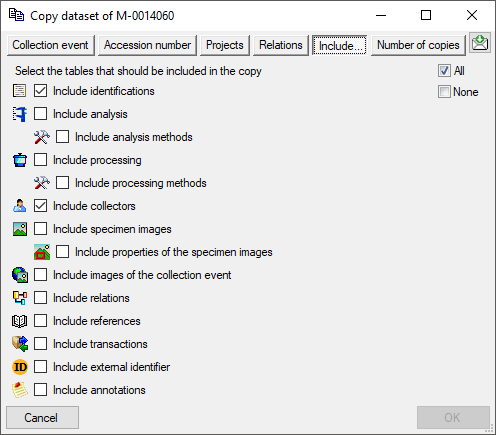
 button to refresh the list.
button to refresh the list.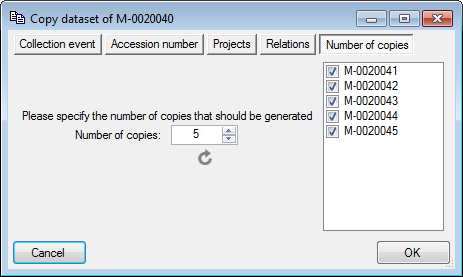
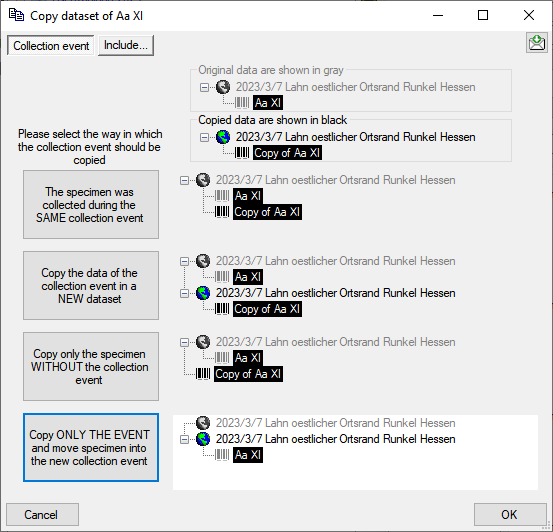
 and the background will turn
yellow to show that there is hidden data of the collectors.
and the background will turn
yellow to show that there is hidden data of the collectors.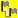 = hide the SamplingPlots
= hide the SamplingPlots . To change to one of these use the
. To change to one of these use the 

 button. The
button. The  button returns it to the list to be shown on a label. The first unit can
not be transferred to the hide list.
button returns it to the list to be shown on a label. The first unit can
not be transferred to the hide list.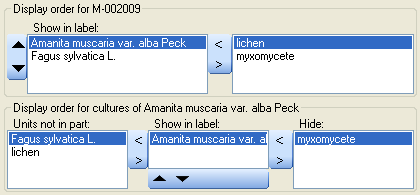
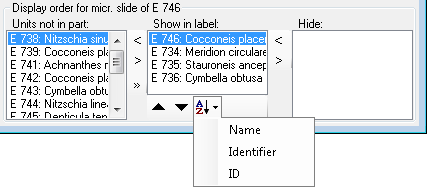
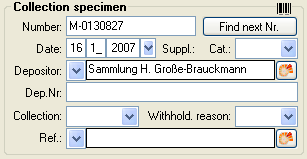

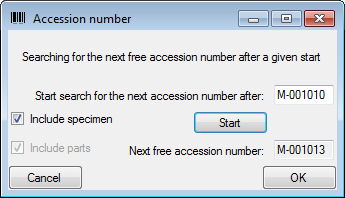

 button: Ordering ascending resp. descending
by the name (= URI) or the creation date of the images. For a short
introduction see the tutorial
button: Ordering ascending resp. descending
by the name (= URI) or the creation date of the images. For a short
introduction see the tutorial
 button
(see below) will change to
button
(see below) will change to 


 .
.
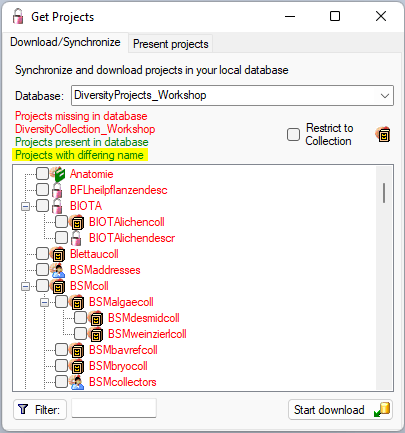
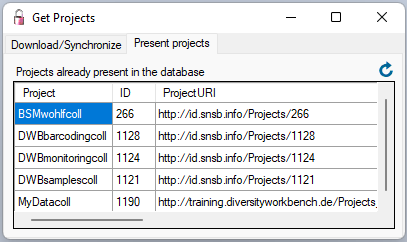

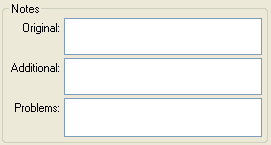
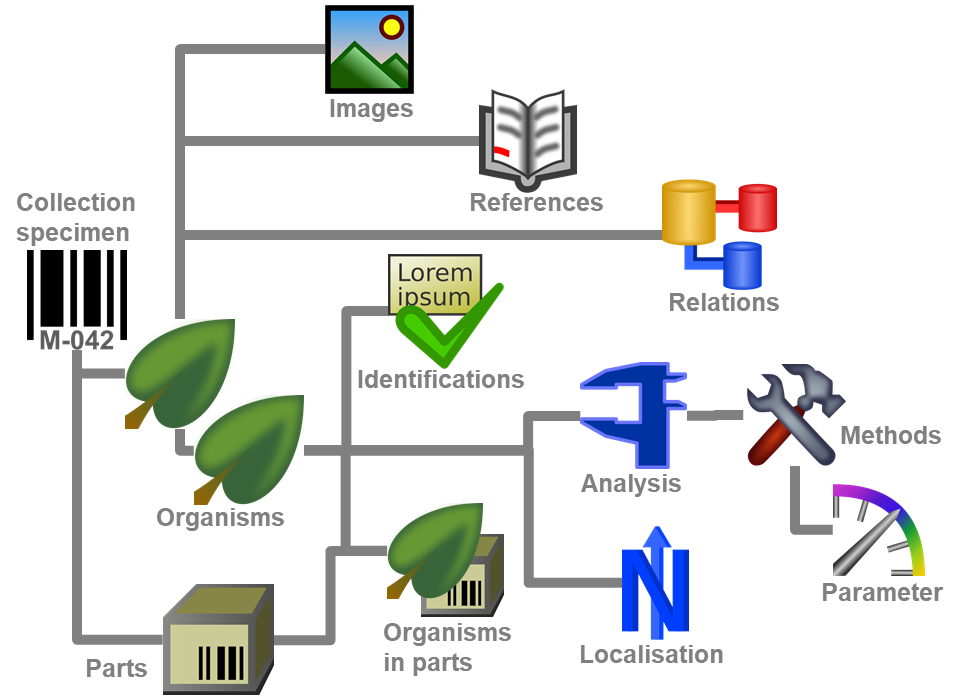



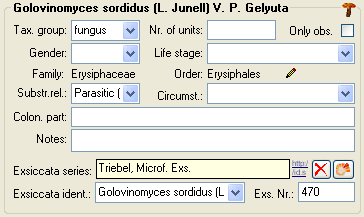



 button next to
the relation type. A window will open where you can choose among the
organisms (= units) in your sample. The second parent will be indicated
with the
button next to
the relation type. A window will open where you can choose among the
organisms (= units) in your sample. The second parent will be indicated
with the 
 and
copy
and
copy  predefined values into the data (see
chapter
predefined values into the data (see
chapter  soil,
soil,
 rock or
rock or 
 button. With the
button. With the 

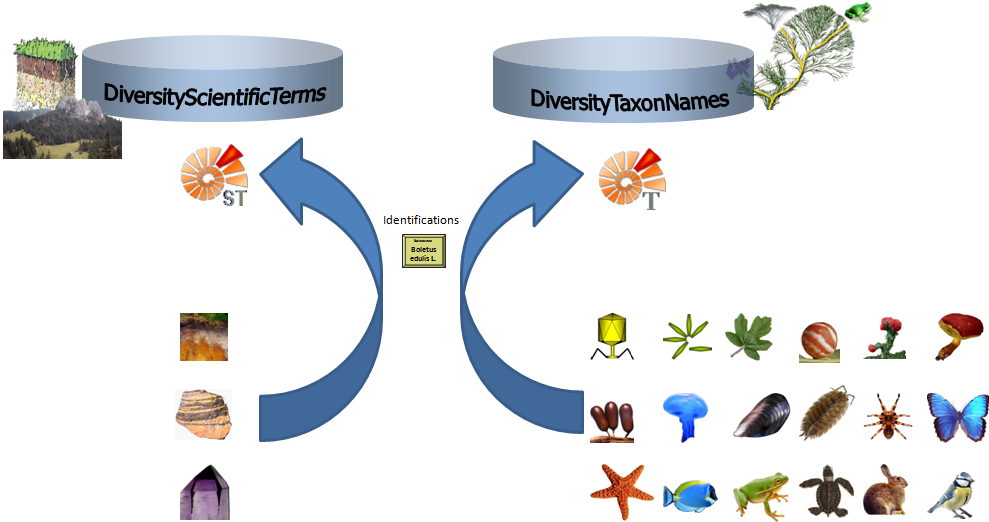

 and the entries will appear as shown
in the tree below. The last state
and the entries will appear as shown
in the tree below. The last state  will hide
the entries.
will hide
the entries.

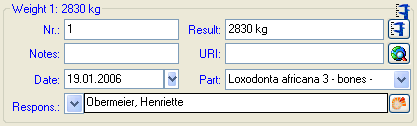

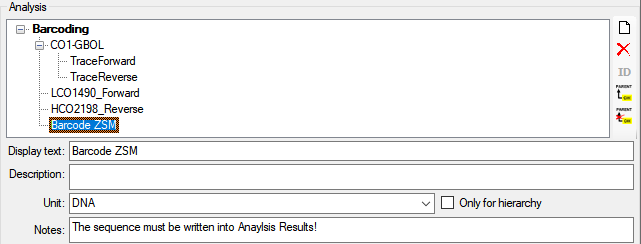

 button opens a separate window
(see below) with additional options for
button opens a separate window
(see below) with additional options for 
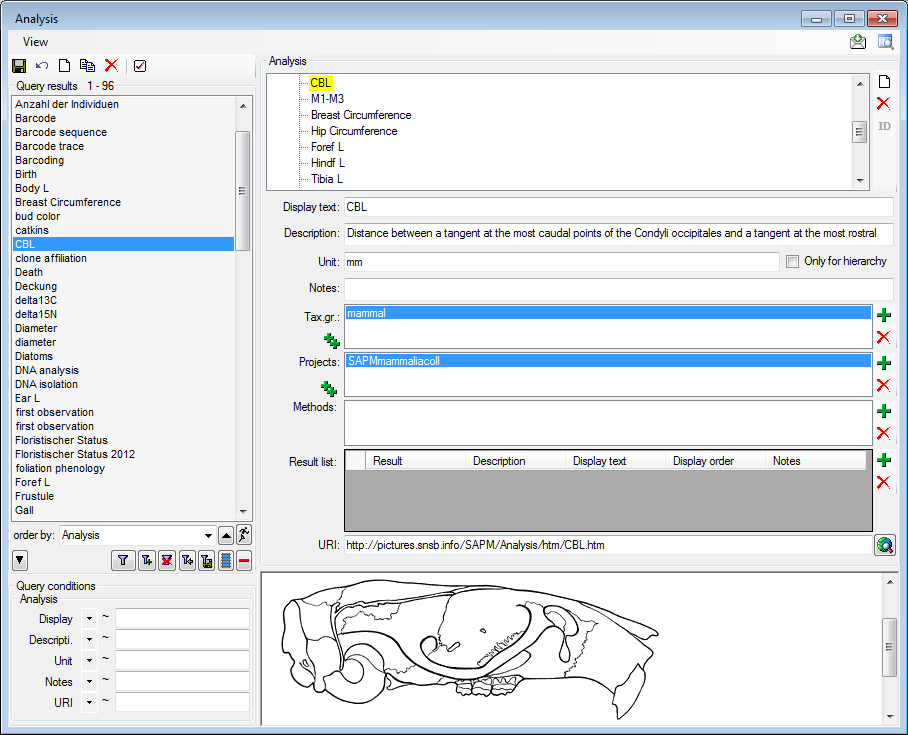

 buttons. For details
about handling data see the
buttons. For details
about handling data see the 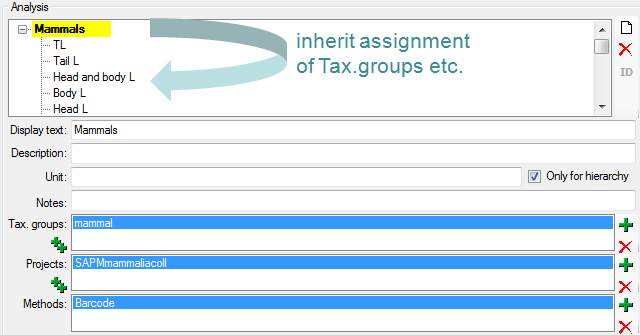
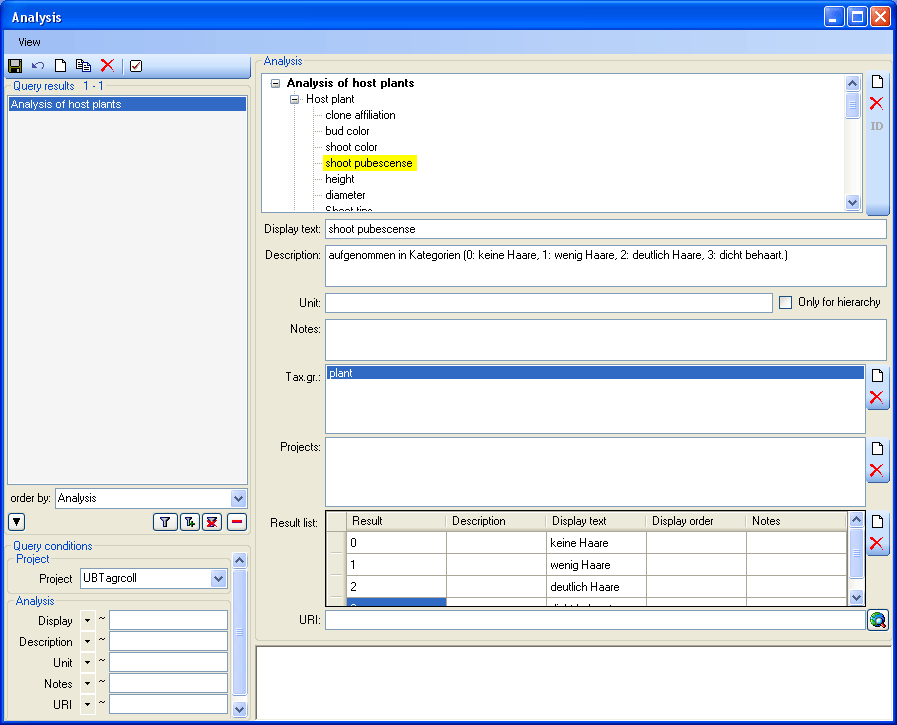

 identification to base icon.
identification to base icon. confirmation button. To move an
identification to the end or the start of the list use the
confirmation button. To move an
identification to the end or the start of the list use the  identification down or
identification down or
 identification up button respectively. To delete an
identification select it in the tree and click the
identification up button respectively. To delete an
identification select it in the tree and click the 



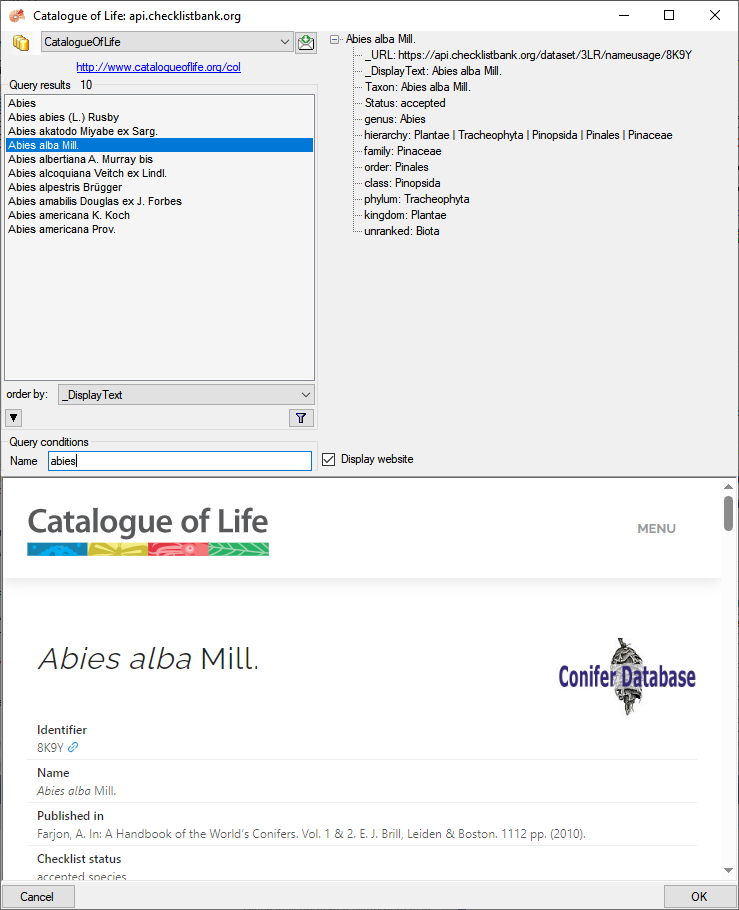

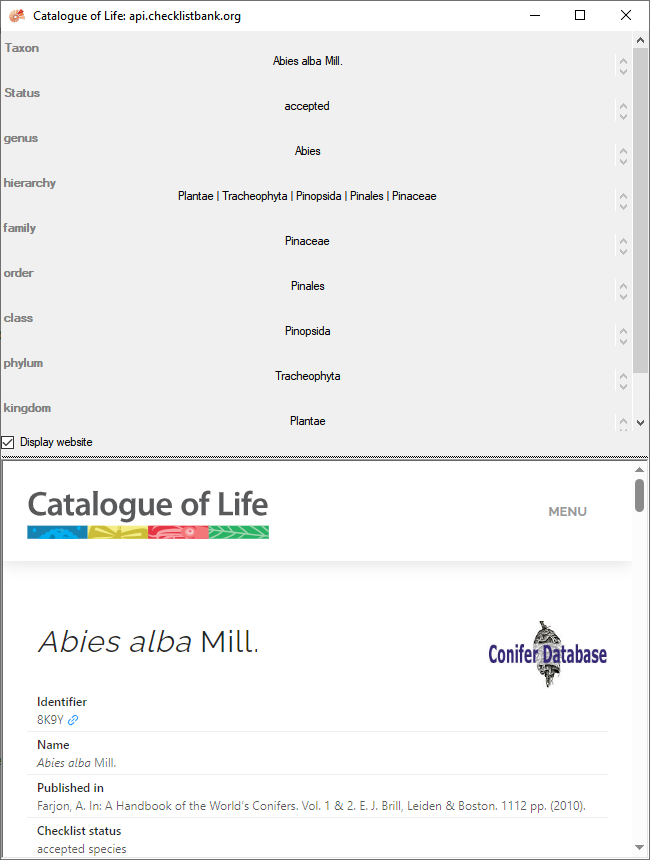
 Index
Fungorum. To establish a connection to this webservice, click on the
Index
Fungorum. To establish a connection to this webservice, click on the
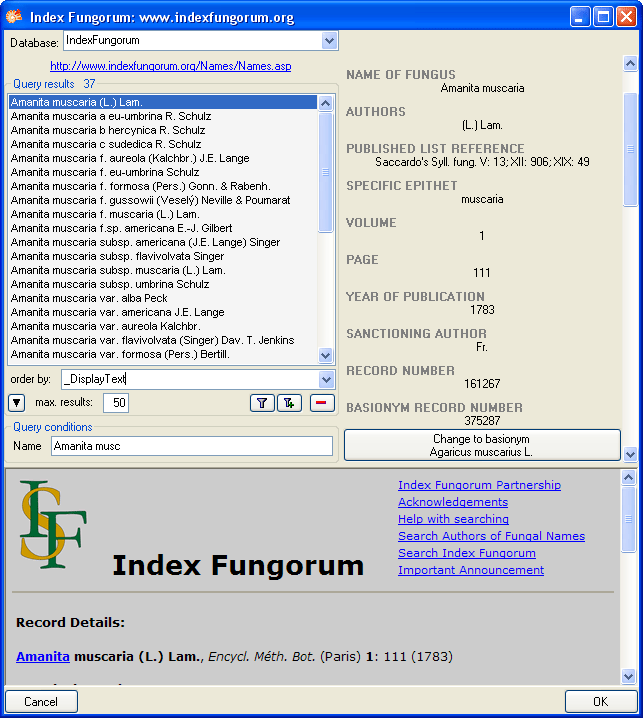
 (choose a low
number, if you have a slow connection to the internet). Click on the
search button
(choose a low
number, if you have a slow connection to the internet). Click on the
search button 
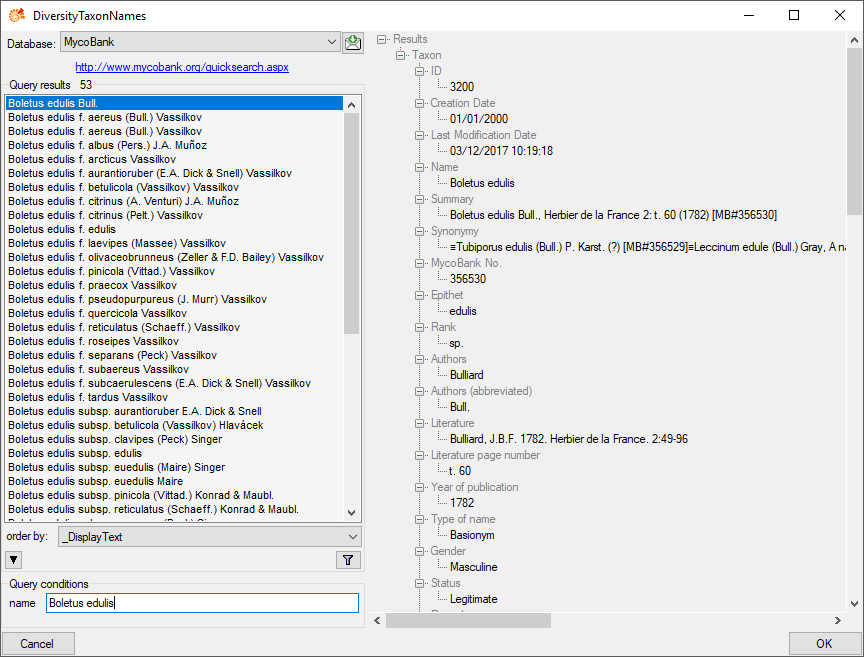
 , a window will open providing you with the retrieval
information of the webservice.
, a window will open providing you with the retrieval
information of the webservice. 
 Pan-European Species directories Infrastructure (PESI). To establish a
connection to this webservice, click on the
Pan-European Species directories Infrastructure (PESI). To establish a
connection to this webservice, click on the



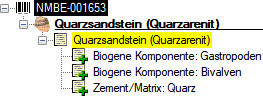
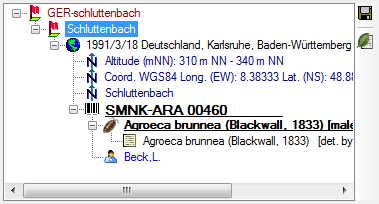
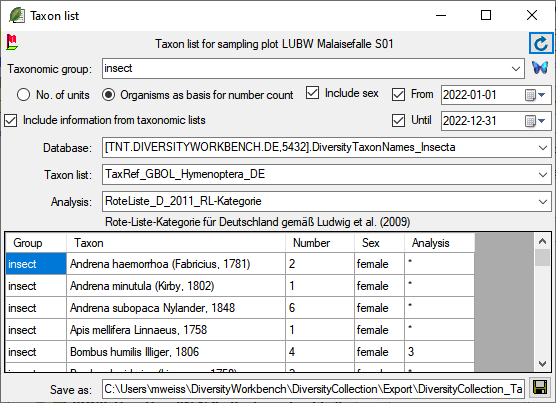
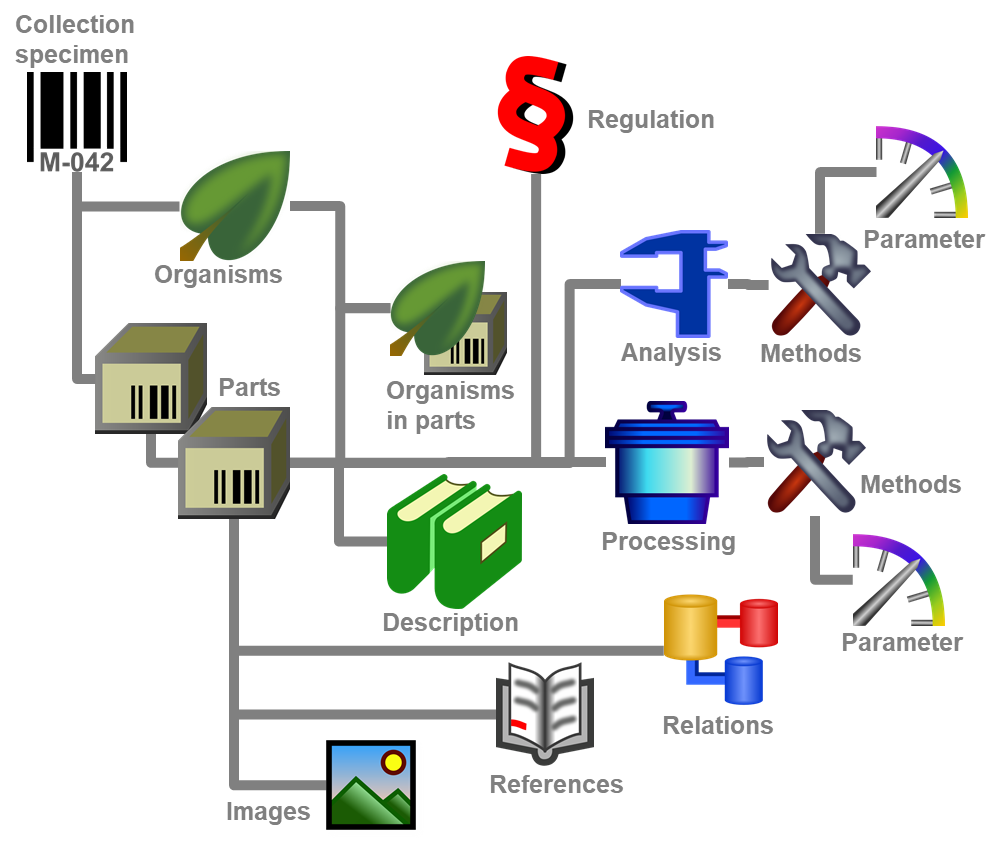
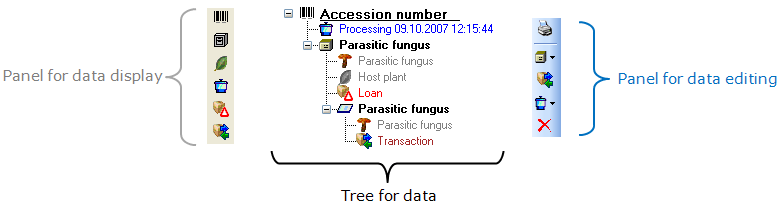
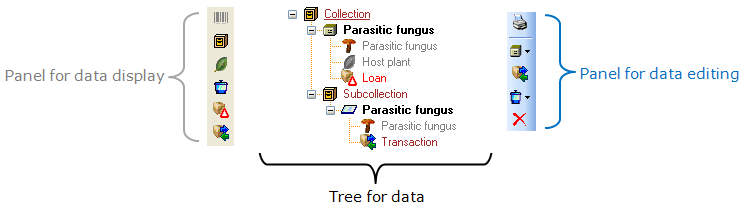
 and the background will turn
yellow, to show you that there is hidden data of the processings.
and the background will turn
yellow, to show you that there is hidden data of the processings.

 button. All specimens within the
collections will be listed as shown below with e.g. their accession
number, accession number of the part, storage location, collection date
and/or locality as set in
button. All specimens within the
collections will be listed as shown below with e.g. their accession
number, accession number of the part, storage location, collection date
and/or locality as set in 
 in the tree. You may enter a
description for an
in the tree. You may enter a
description for an  icon, was created from the sample. Thus, in the
specimen tree it is shown as a child of the specimen symbolized by the
icon, was created from the sample. Thus, in the
specimen tree it is shown as a child of the specimen symbolized by the
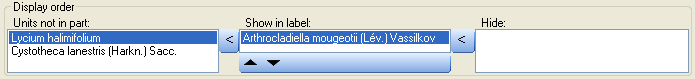
 . A window will open listing all previous volumns
together unit, the date and the responsible user.
. A window will open listing all previous volumns
together unit, the date and the responsible user.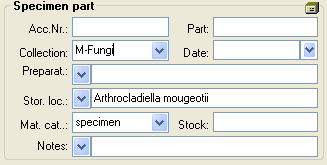
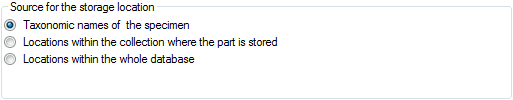
 bones: bones or skeleton from vertebrates
bones: bones or skeleton from vertebrates cultures: living cultures of organisms
cultures: living cultures of organisms drawing: original line or color drawing
drawing: original line or color drawing herbarium sheets: capsules or sheets as
stored in a botanical collection
herbarium sheets: capsules or sheets as
stored in a botanical collection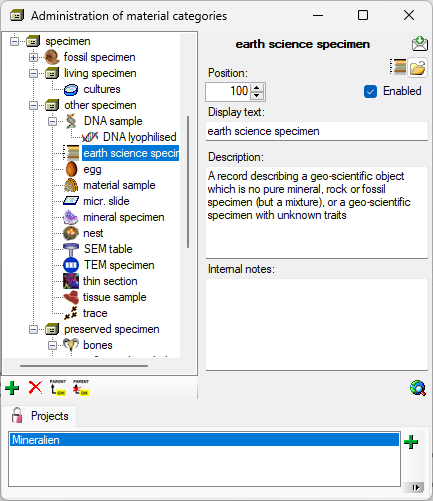
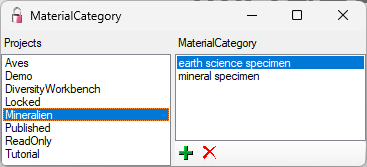

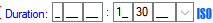
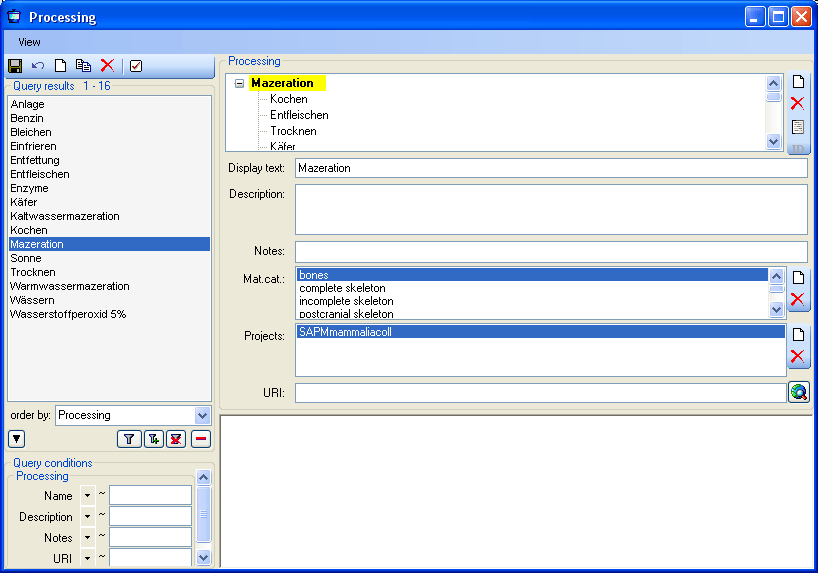
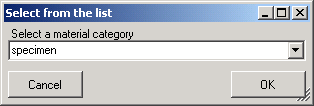



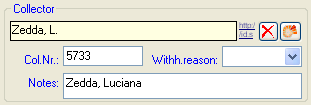
 ,
,  ), identification
), identification
 ) in the tree and click on the
) in the tree and click on the 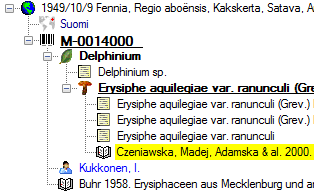
 ) or part (e.g.
) or part (e.g.
 buttons. For an overview see a short
tutorial
buttons. For an overview see a short
tutorial
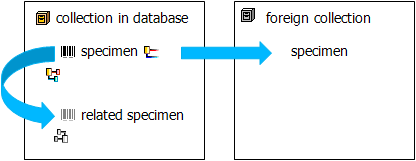
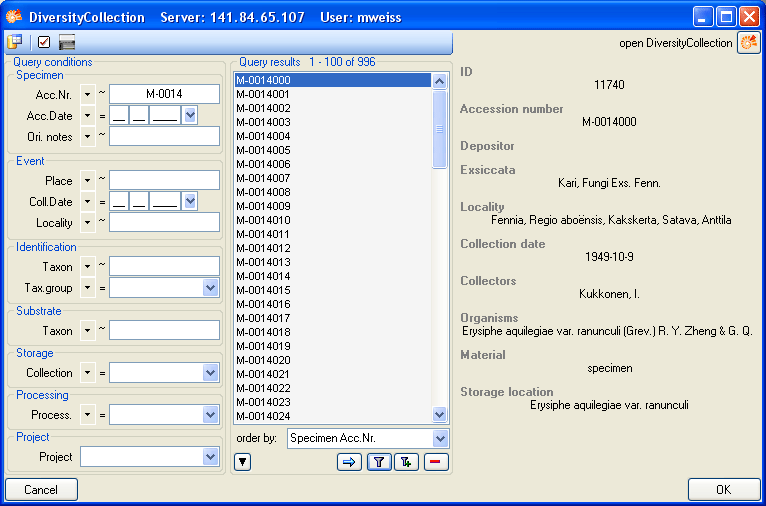
 from another specimen.
from another specimen. 


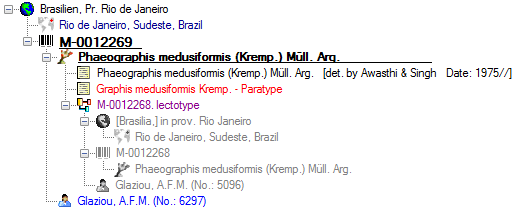


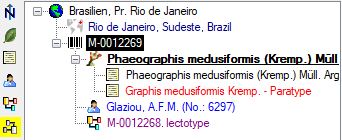

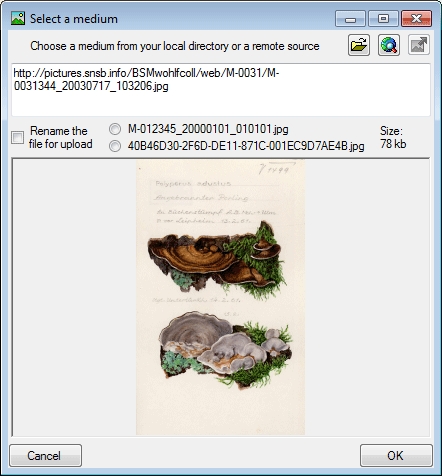
 upload button to upload your image to the server.
upload button to upload your image to the server.
 will appear as shown below.
will appear as shown below.



 and video
and video

 button. The annotation will be
shown in the tree with the details listed below (see image below).
button. The annotation will be
shown in the tree with the details listed below (see image below).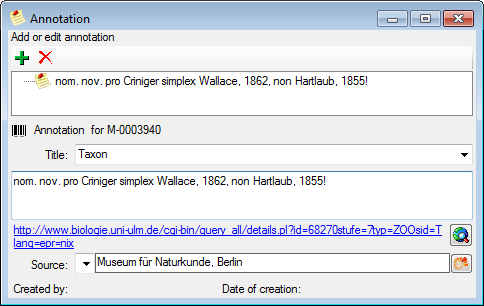
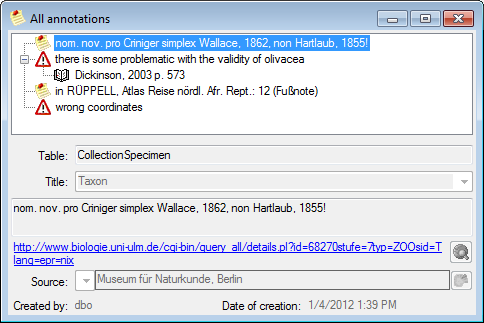
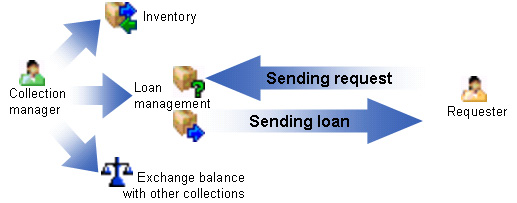


 Superior datasets and children of
current dataset
Superior datasets and children of
current dataset Restrict to superior datasets
Restrict to superior datasets Hide the hierarchy
Hide the hierarchy

 button. The latter will copy the collection including all child
collections. To set the superior collection, use the
button. The latter will copy the collection including all child
collections. To set the superior collection, use the 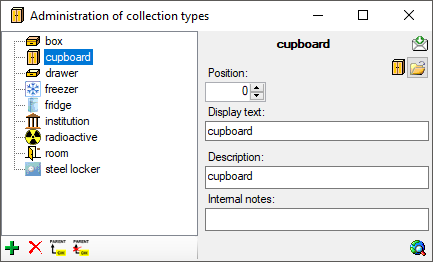


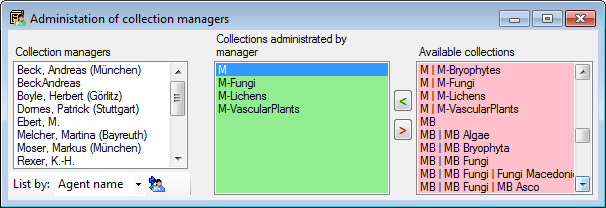
 agent synchonisation button to correct this. In case of
duplicate permissions you might get a list with these duplicates where
you have to remove the duplicate permissions.
agent synchonisation button to correct this. In case of
duplicate permissions you might get a list with these duplicates where
you have to remove the duplicate permissions.
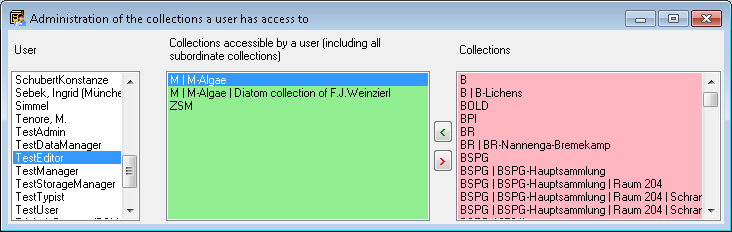
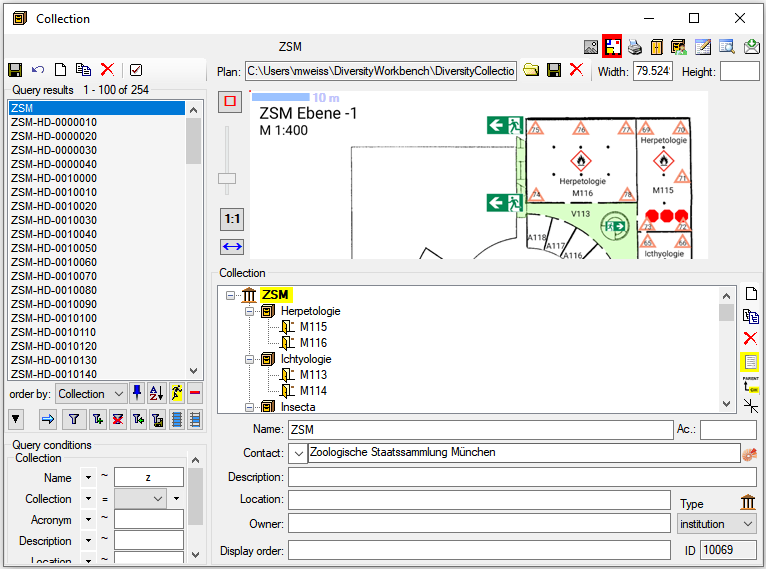
 (see below). The administrative hierarchy is ignored concerning the inheritance of floor plan and geometries.
(see below). The administrative hierarchy is ignored concerning the inheritance of floor plan and geometries.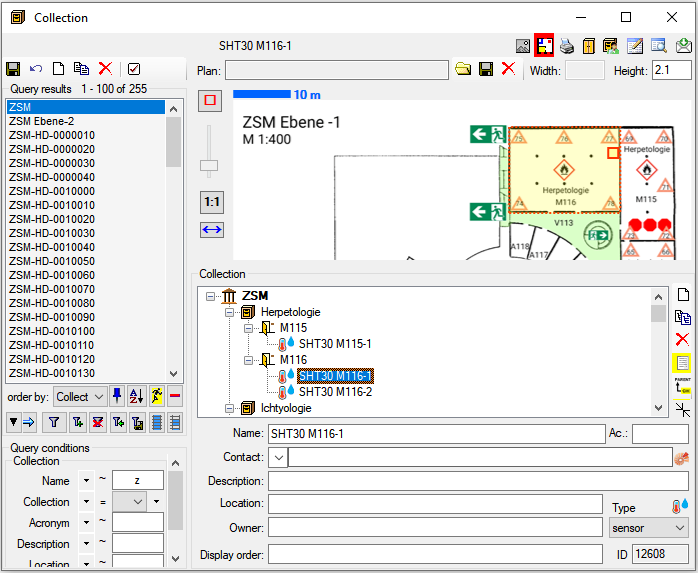
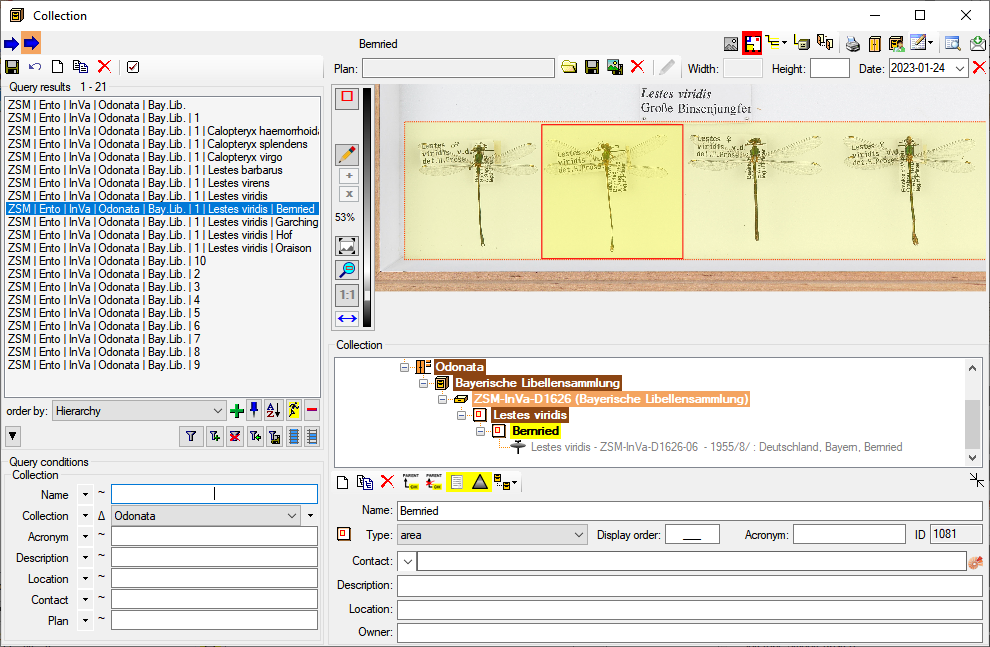

 Defaults and miscellaneous
Defaults and miscellaneous Number of displayed levels for location hierarchy
Number of displayed levels for location hierarchy
 button to zoom in the geometry.
With the
button to zoom in the geometry.
With the 


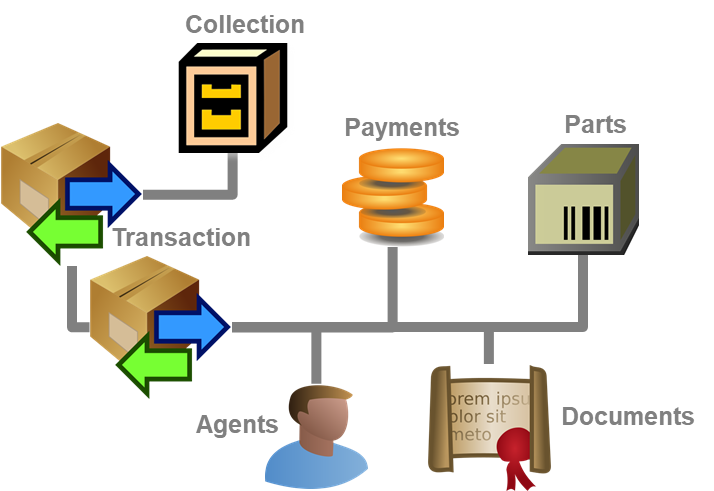
 icon (see image below). A
icon (see image below). A
 icon. If there are older accession numbers
linked to a specimen involved in a transaction, these numbers can be
documented together with the entry of the transaction of the respective
part of the specimen.
icon. If there are older accession numbers
linked to a specimen involved in a transaction, these numbers can be
documented together with the entry of the transaction of the respective
part of the specimen.
 botton resp.
botton resp.

 gift
gift inventory
inventory permanent loan
permanent loan
 purchase
purchase

 transaction group
transaction group
 button to
create a document. To print the document use the
button to
create a document. To print the document use the 
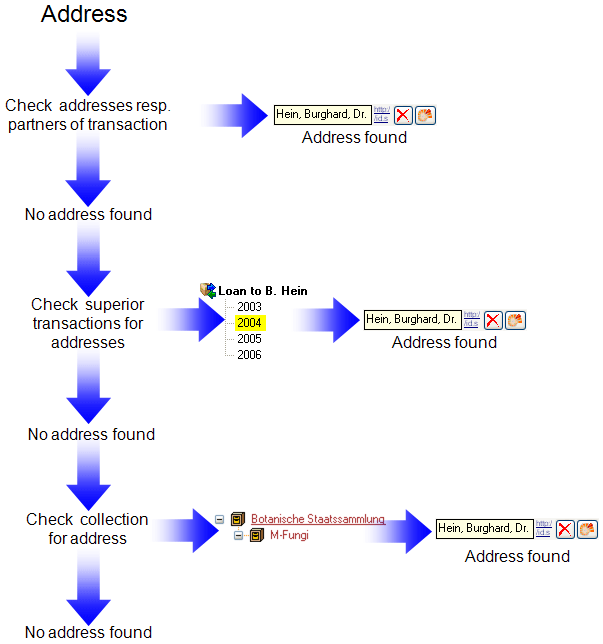
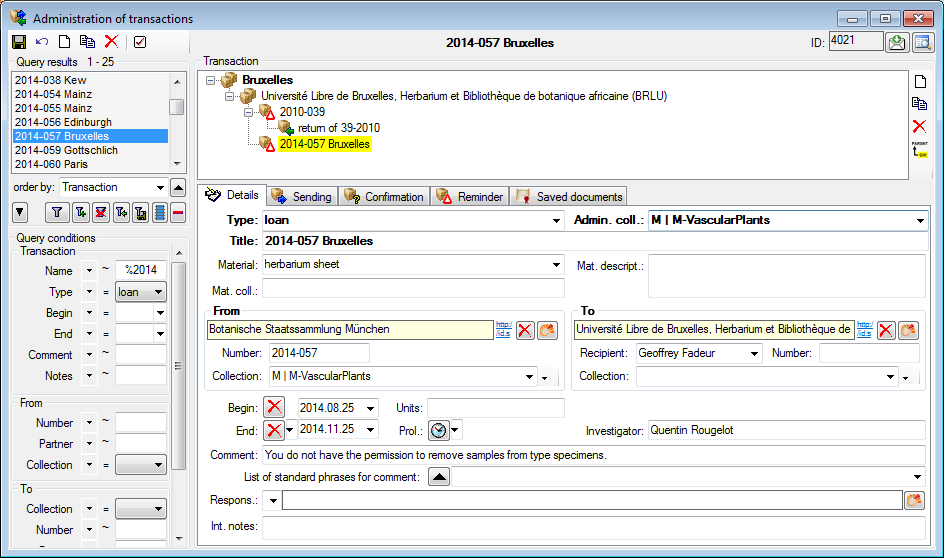




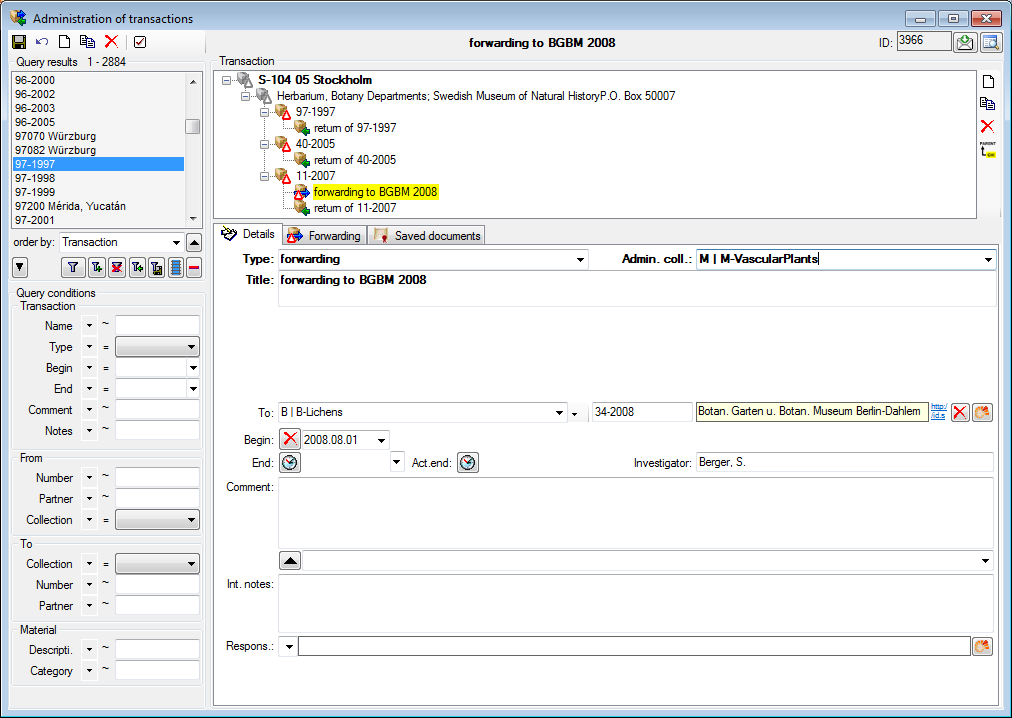

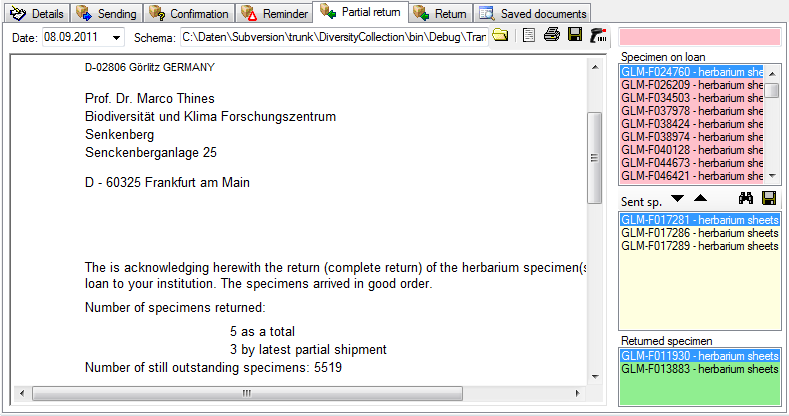

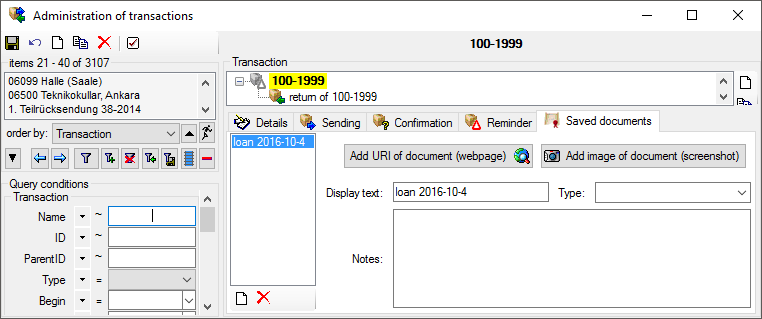
 Add image of document.
The first option uses public available sources from a webserver while
with the second option later screenshots will be stored directly in the
database and are not accessible outside the database. If you want to
print the document, double click on the image to open a window enabling
the printing of the document
Add image of document.
The first option uses public available sources from a webserver while
with the second option later screenshots will be stored directly in the
database and are not accessible outside the database. If you want to
print the document, double click on the image to open a window enabling
the printing of the document  “https://…”
button to search for an
URL in your default browser instead of the inbuilt browser of the
software which may not provide the whole functionality of a modern
browser.
“https://…”
button to search for an
URL in your default browser instead of the inbuilt browser of the
software which may not provide the whole functionality of a modern
browser. option and use the
option and use the
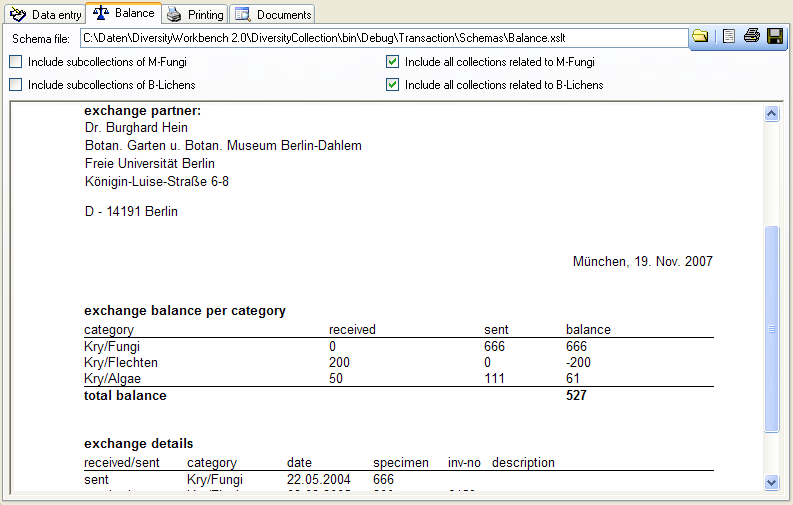
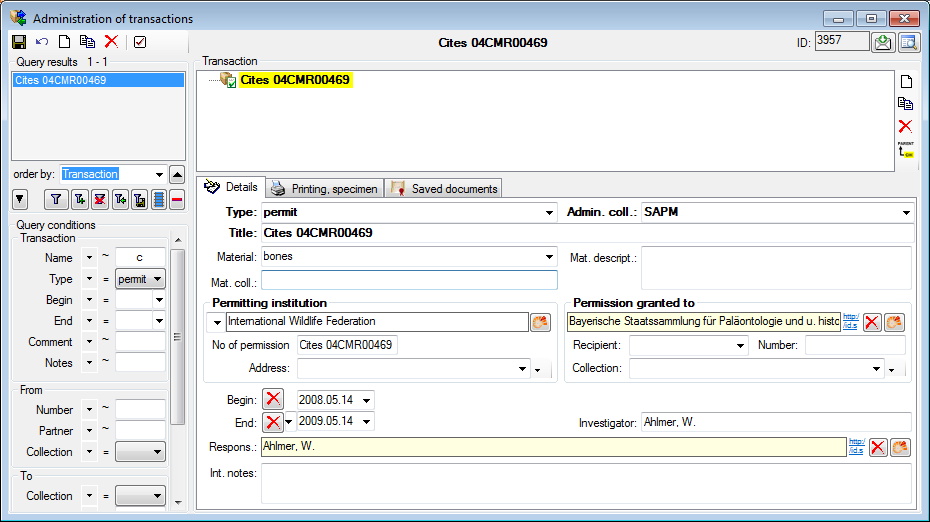
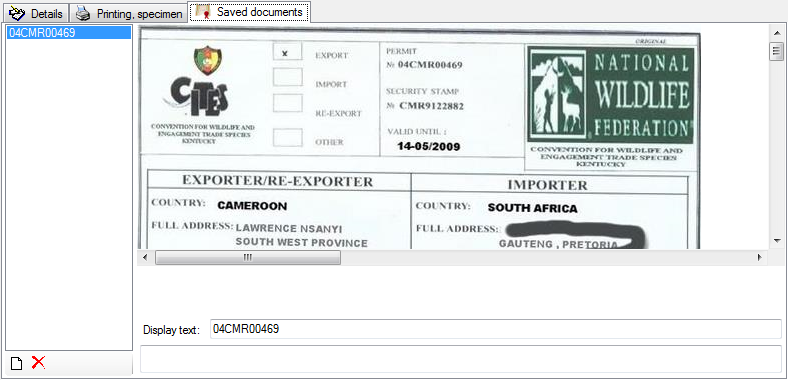
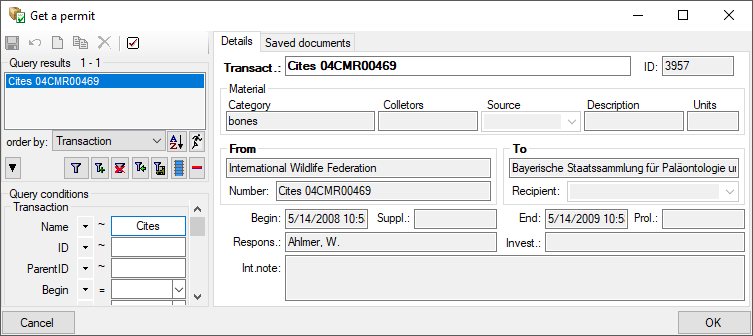
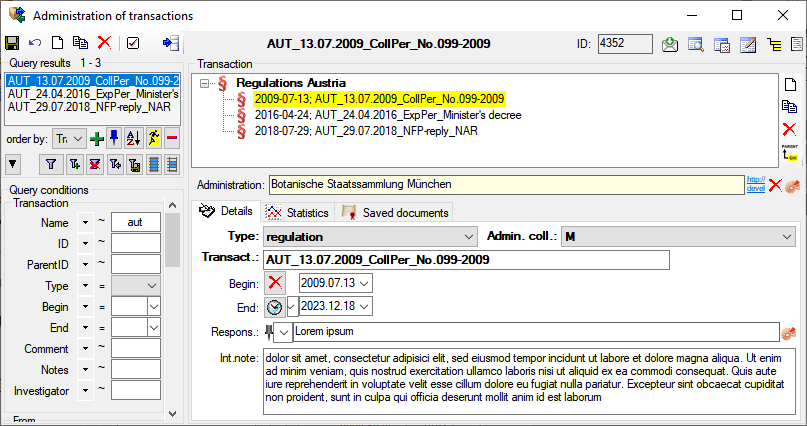
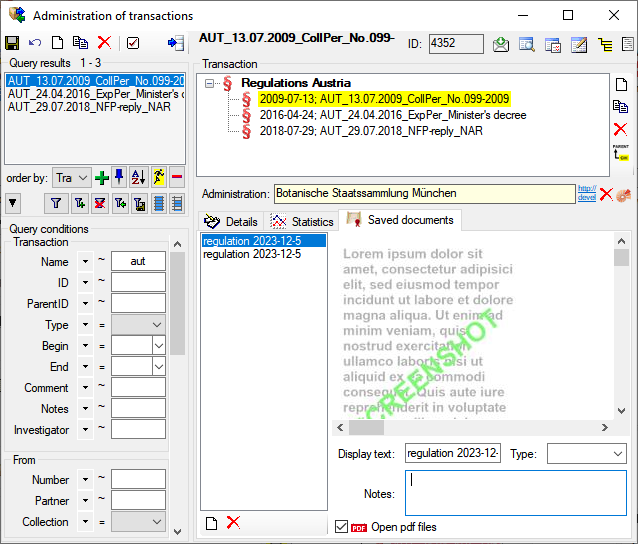

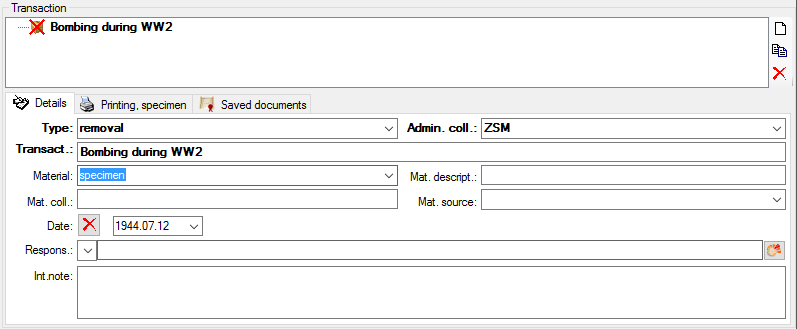
 .
.

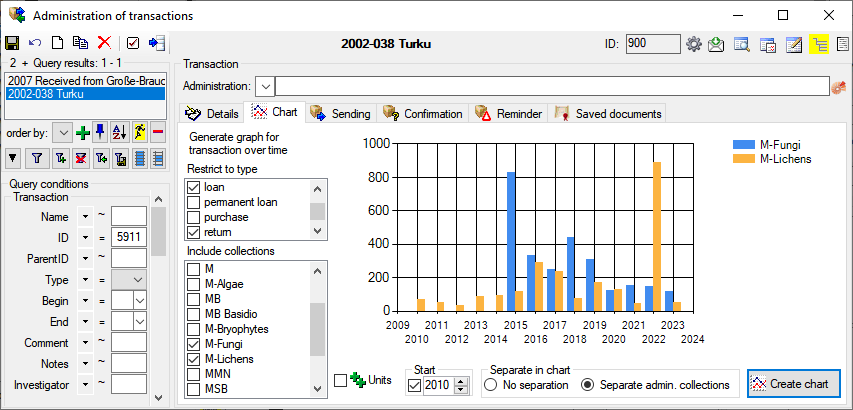

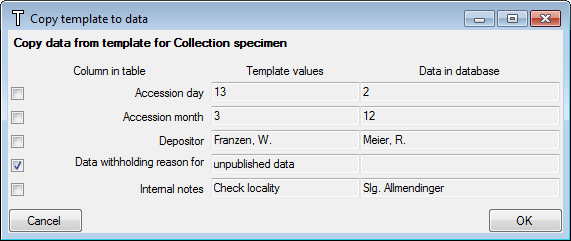
 TaskResult and
TaskResult and 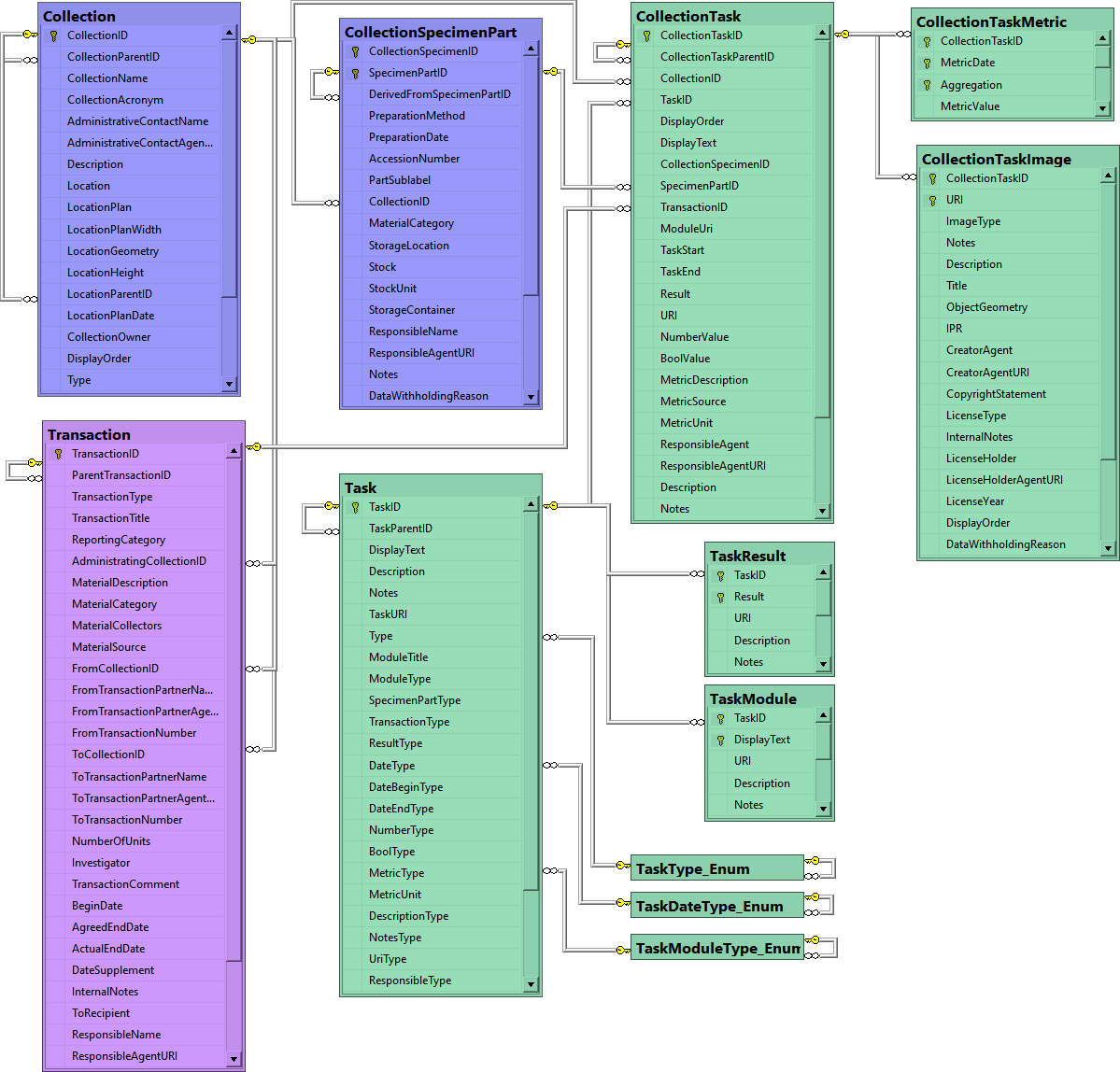
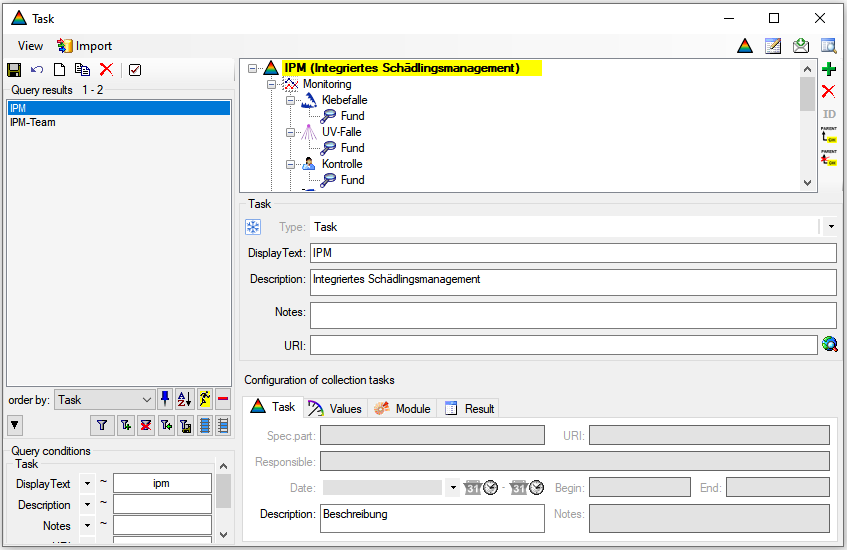
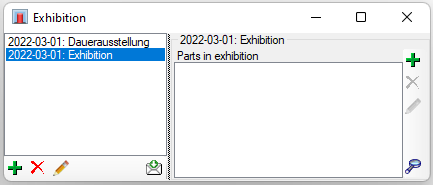
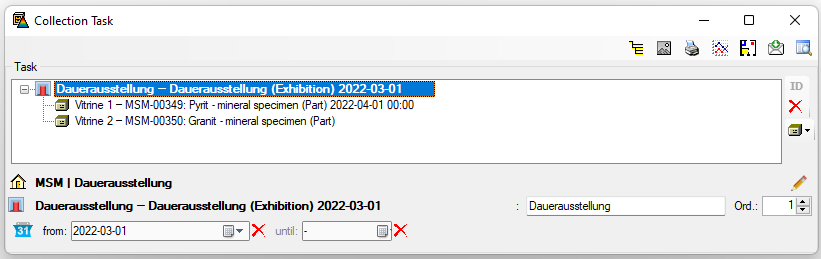
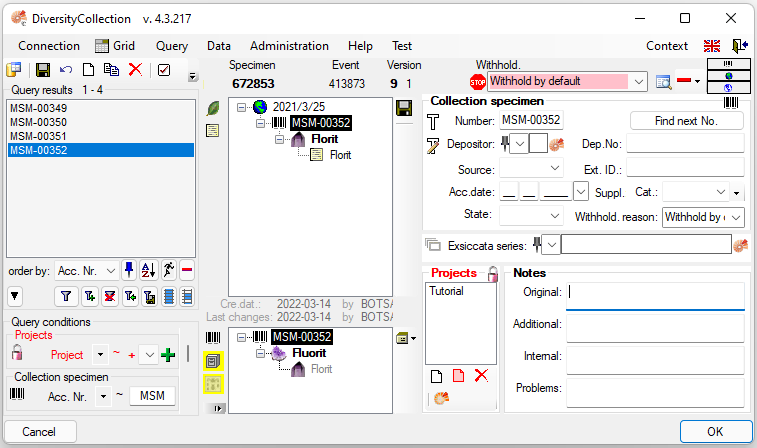
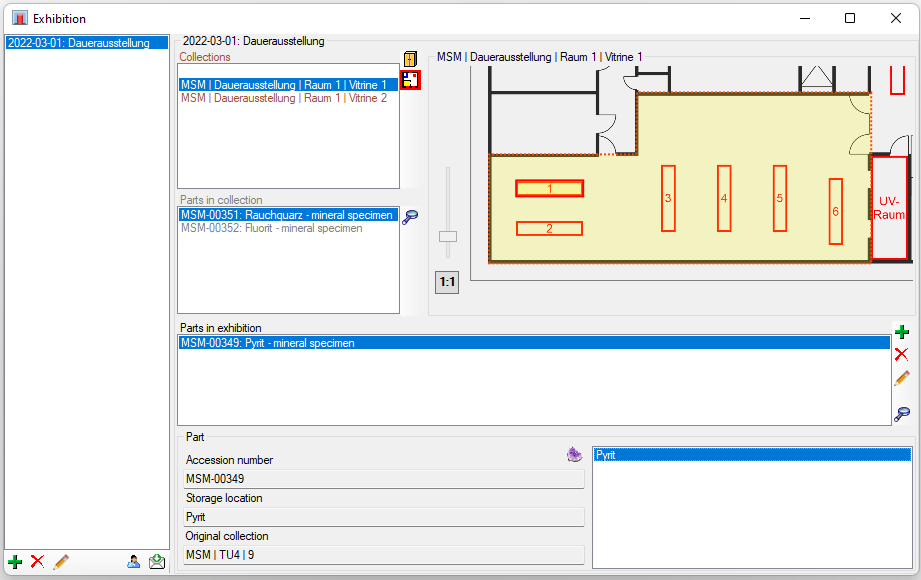
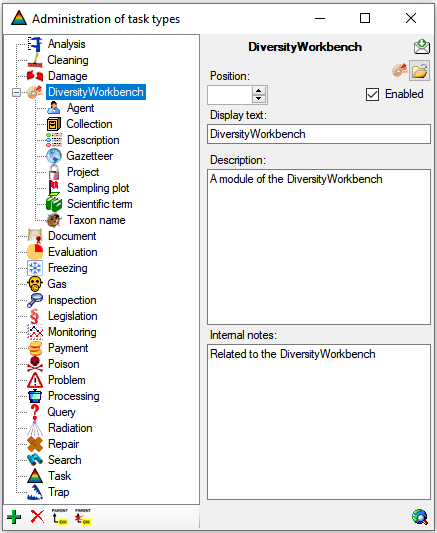

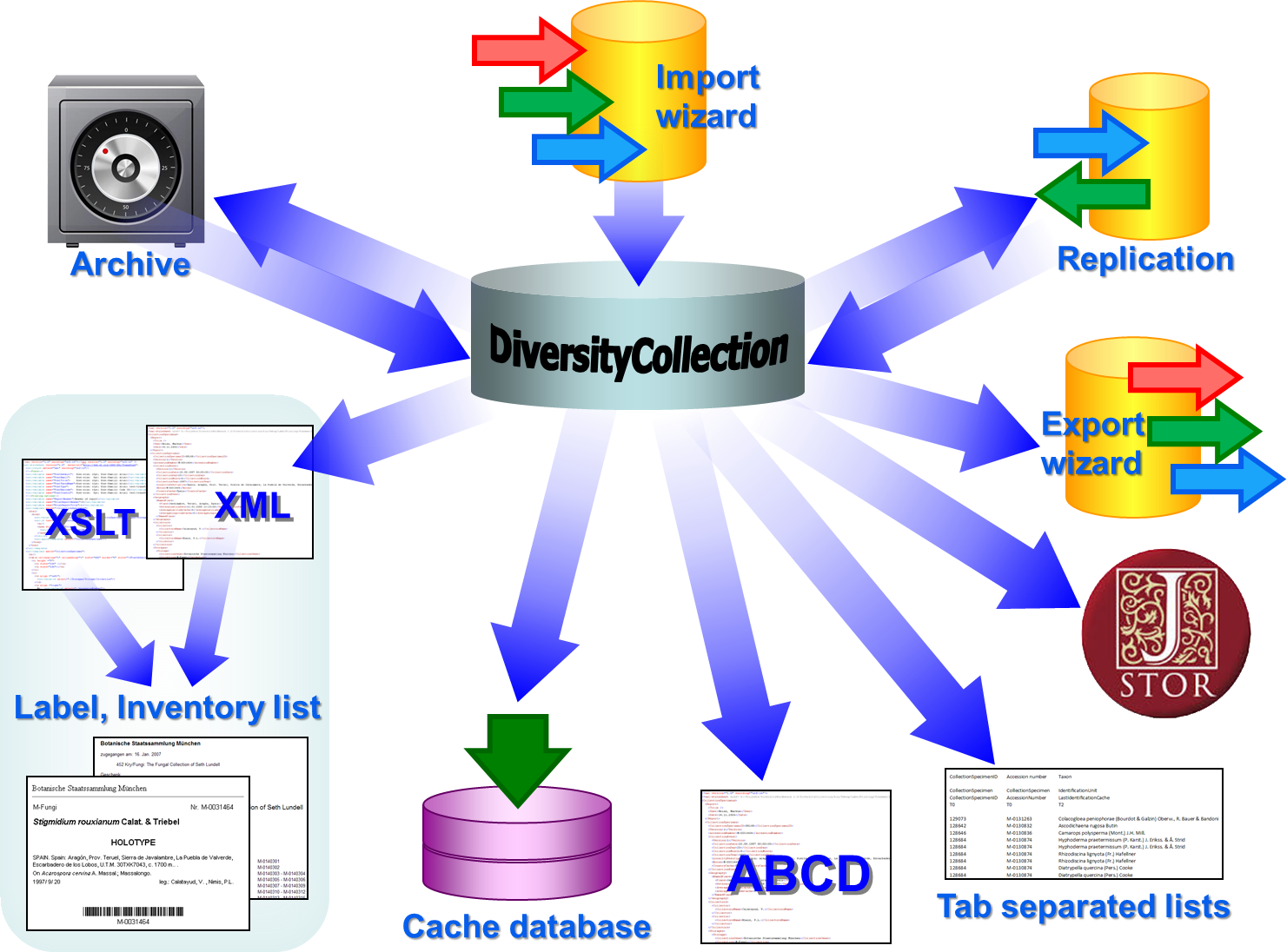
 button. The data related
with the project will be imported into temporary tables to allow you to
inspect them in advance of the creation of the archive (use the
button. The data related
with the project will be imported into temporary tables to allow you to
inspect them in advance of the creation of the archive (use the 
 button to import the data from the XML files
into temporary tables.
button to import the data from the XML files
into temporary tables.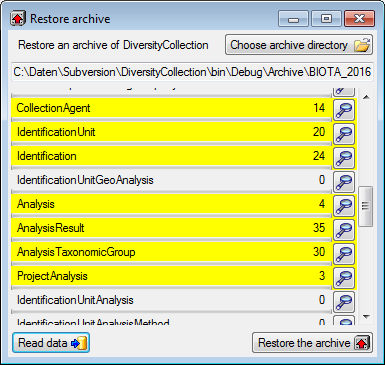
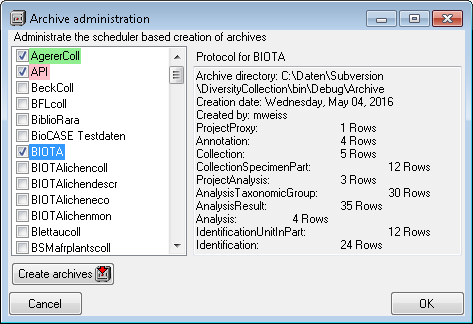
 database
database



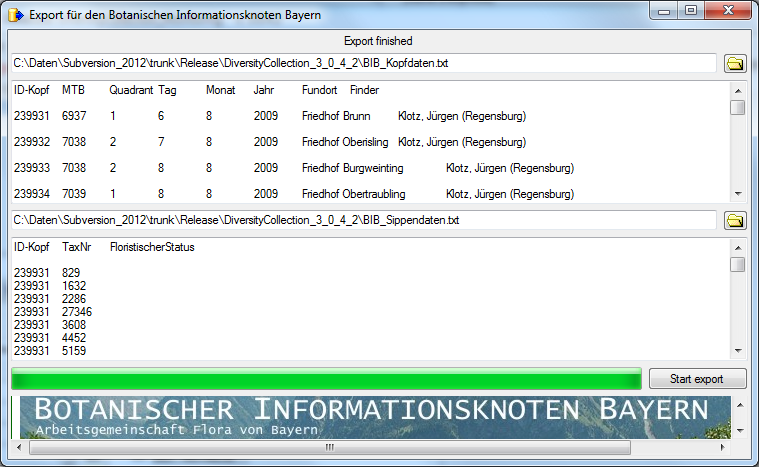
 Several parallel tables according to
selected data
Several parallel tables according to
selected data Dependent table
Dependent table resp.
resp.  button. To fuse
a column with the previous column, click in the gray bar
button. To fuse
a column with the previous column, click in the gray bar
 on the left side of the column that will change to
on the left side of the column that will change to  for fused columns. To remove a file column, use the
for fused columns. To remove a file column, use the  button.
button.
 This filter in contrast to the filter above strictly applies to the row
according to the sequence of the data. For an explanation see a short
tutorial
This filter in contrast to the filter above strictly applies to the row
according to the sequence of the data. For an explanation see a short
tutorial
 To test the export choose the
To test the export choose the 
 Seq starting at the first position and
Seq starting at the first position and
 starting at the last position. Click on
the button Test the transformation to see the result of your
transformation.
starting at the last position. Click on
the button Test the transformation to see the result of your
transformation.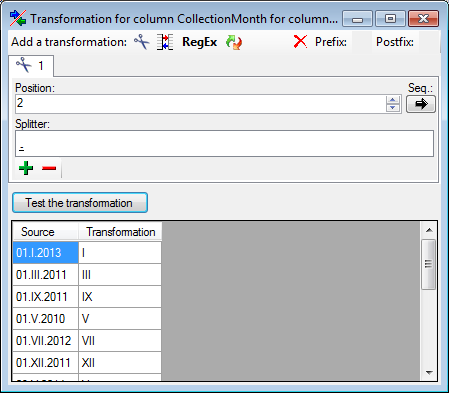
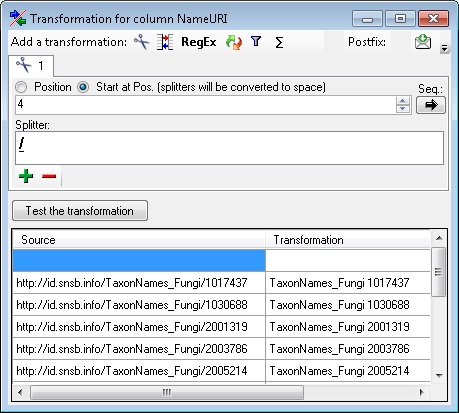
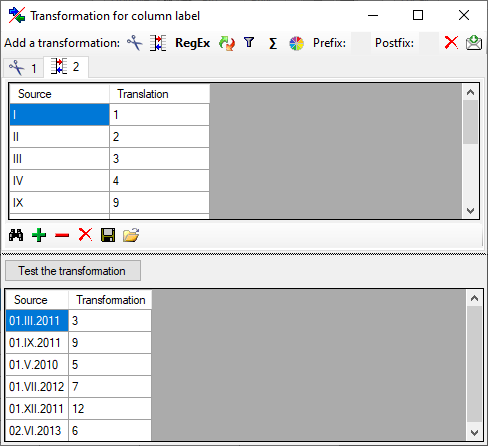
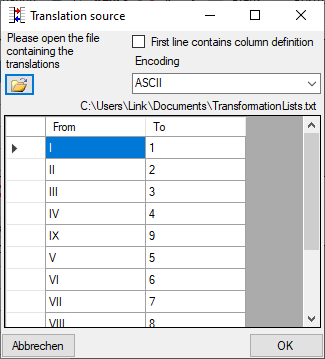
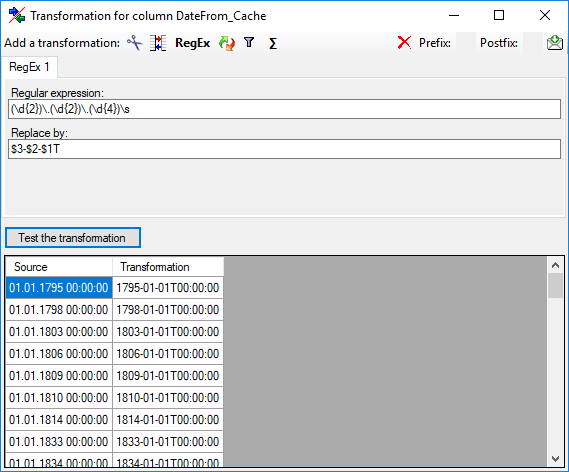
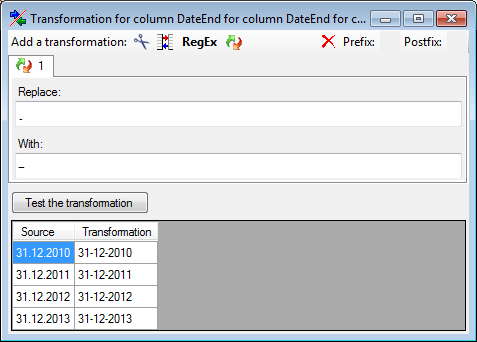
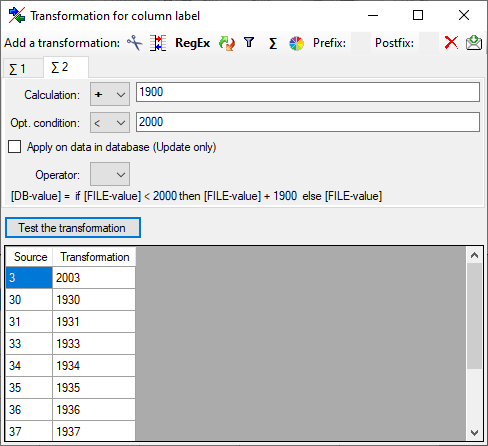
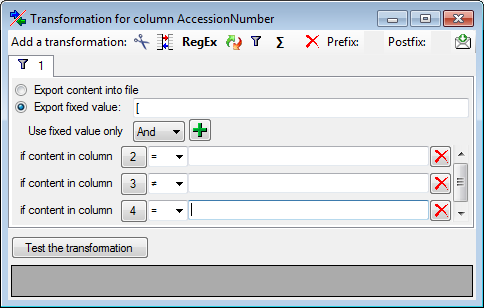
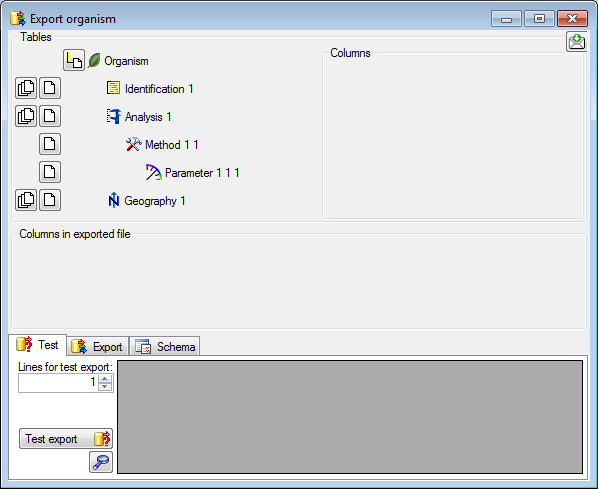
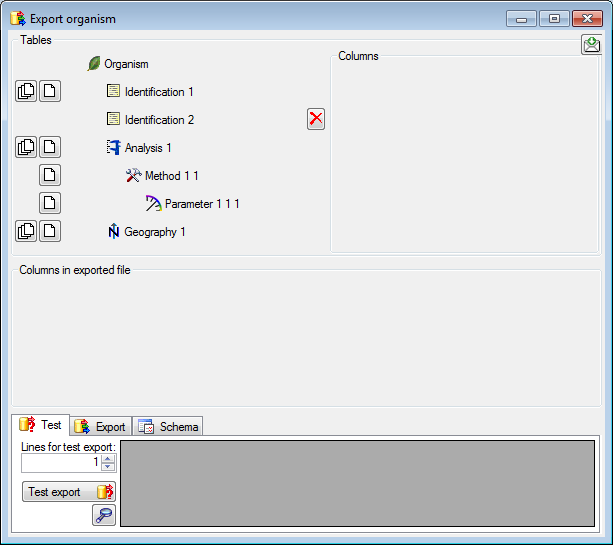
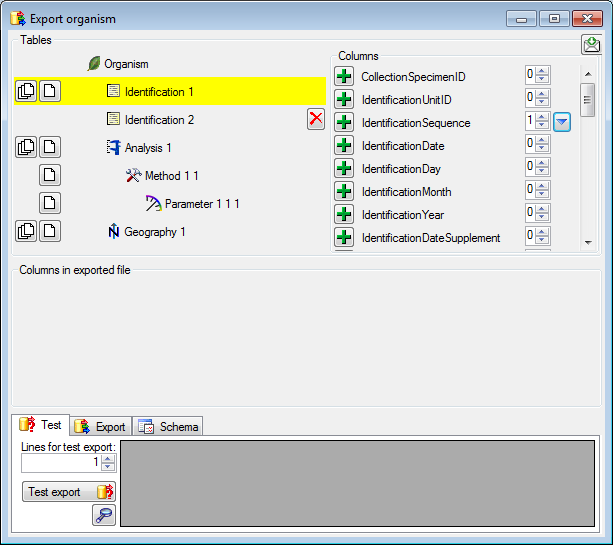
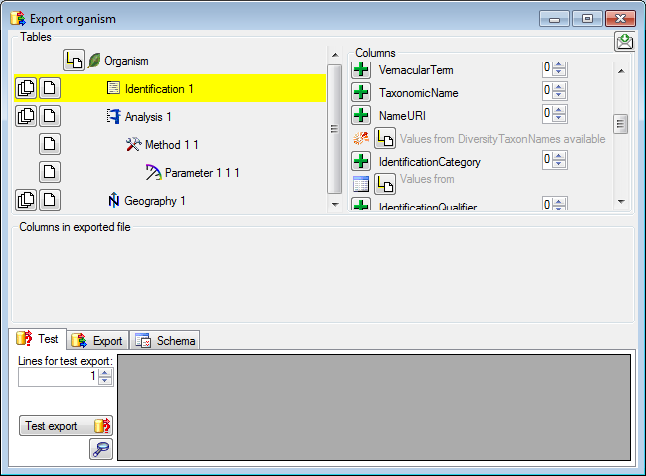
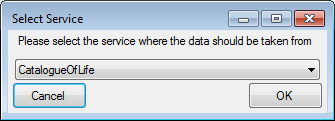
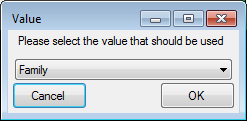
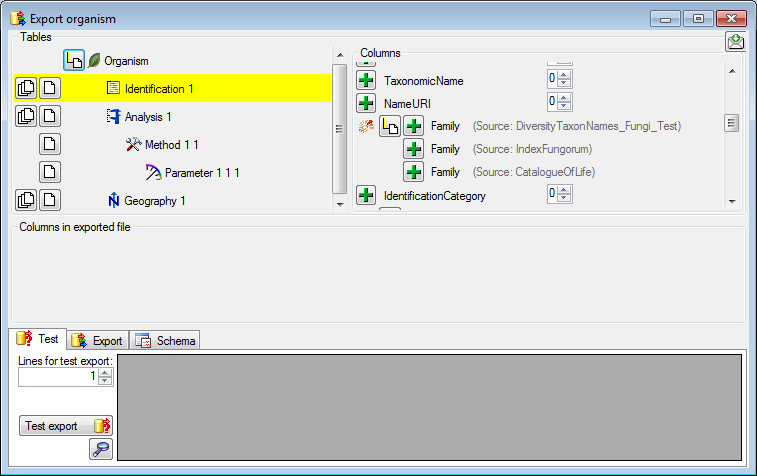
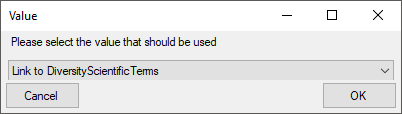


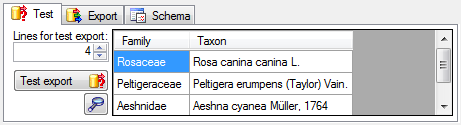
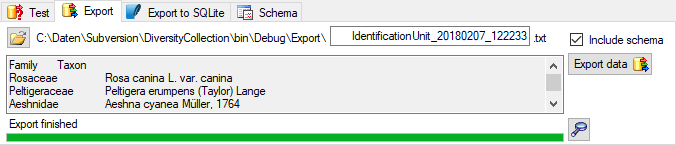

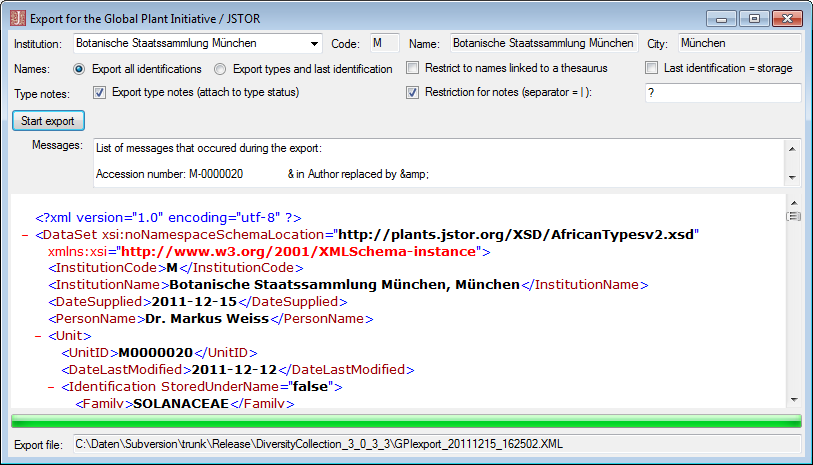
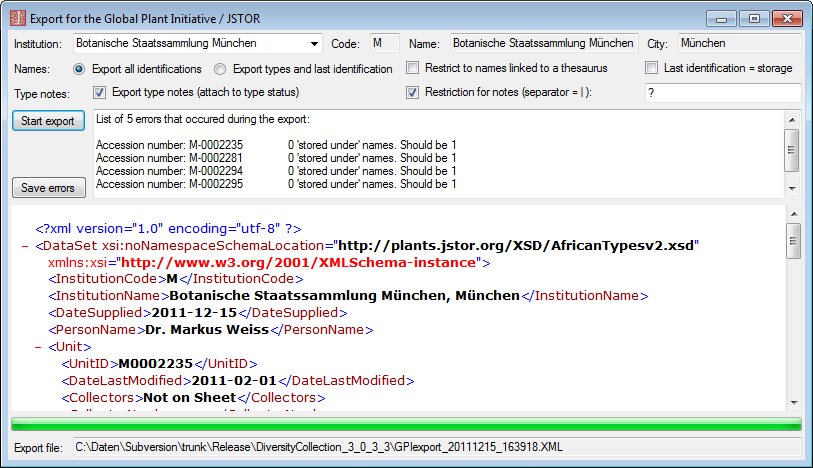
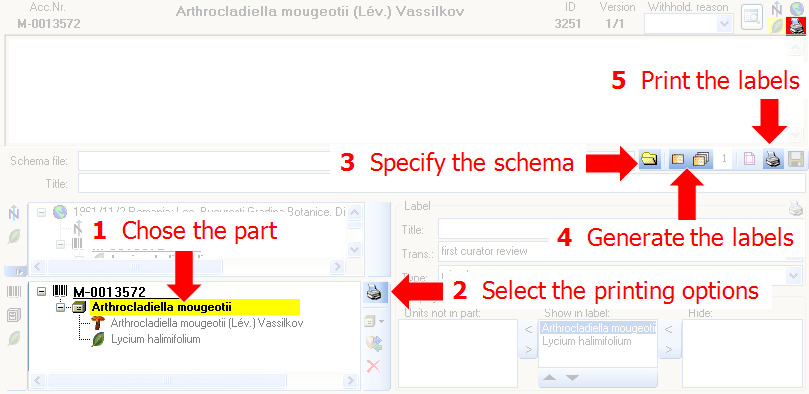
 button (you may use the
button (you may use the
 to the desired value. You can print 1 - 99
duplicates of one label. If there are more than 20 specimens in the
list, you receive a warning whether you really wish to create all these
labels as this could be somewhat time consuming. The labels are
generated as XML files with XSLT-schema files, transformed to HTML-files
and depicted in a browser. To print the label click on
the
to the desired value. You can print 1 - 99
duplicates of one label. If there are more than 20 specimens in the
list, you receive a warning whether you really wish to create all these
labels as this could be somewhat time consuming. The labels are
generated as XML files with XSLT-schema files, transformed to HTML-files
and depicted in a browser. To print the label click on
the 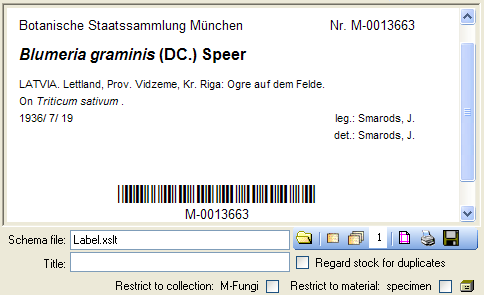
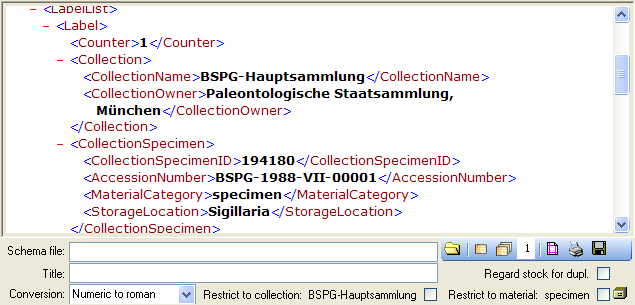



 , which is included in
the DiversityCollection package. Place this font in the folder where
your fonts are stored (e.g.: C:\WINNT\Fonts). If the font is not
available, the barcode will appear as the accession number between two
'*' signs. If this font does not do the job you may try other fonts,
e.g.
, which is included in
the DiversityCollection package. Place this font in the folder where
your fonts are stored (e.g.: C:\WINNT\Fonts). If the font is not
available, the barcode will appear as the accession number between two
'*' signs. If this font does not do the job you may try other fonts,
e.g. 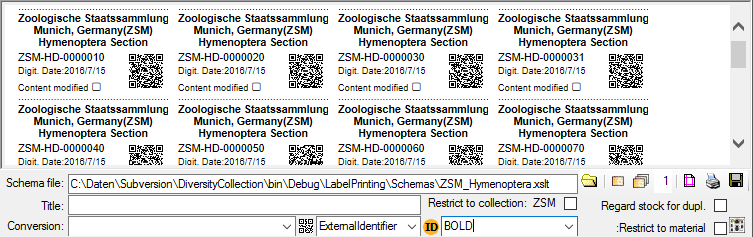

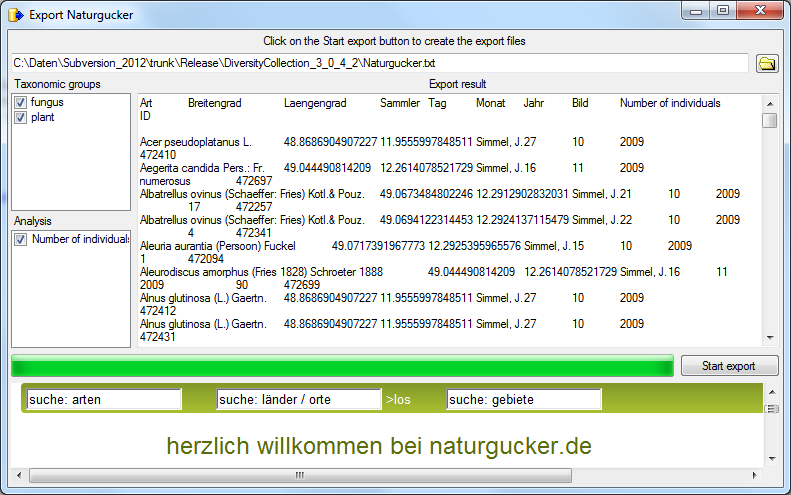
 Postgres (Version 9.4 or above) cache
databases located on any Windows or Linux server (see image below). An
overview for the basic steps is provided in chapter
Postgres (Version 9.4 or above) cache
databases located on any Windows or Linux server (see image below). An
overview for the basic steps is provided in chapter 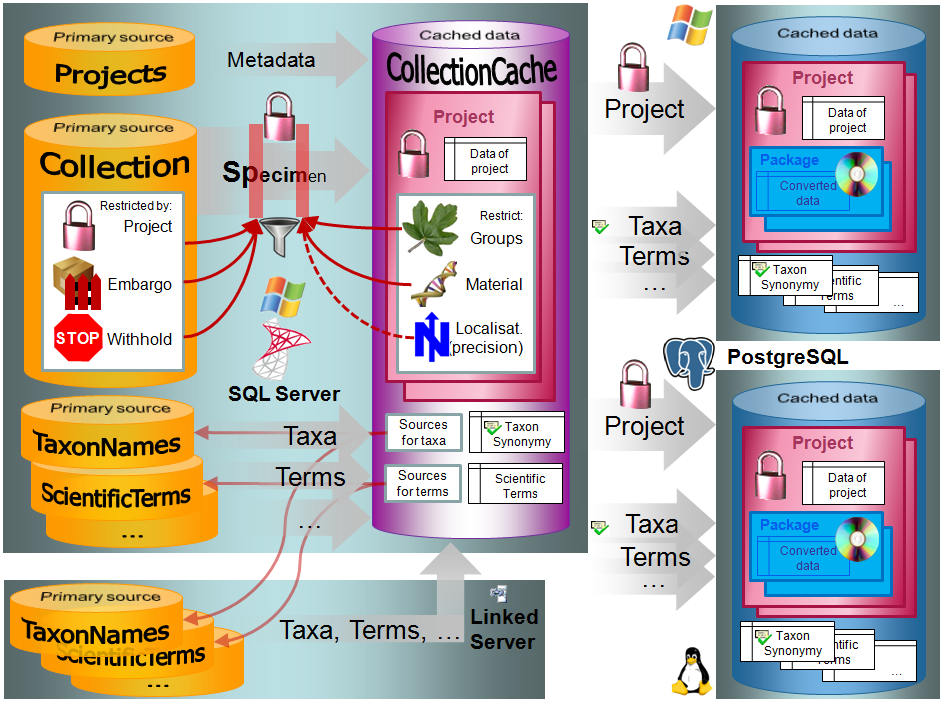
 and synonyms are
transferred into the cache database and may be retrieved from the local
server or a
and synonyms are
transferred into the cache database and may be retrieved from the local
server or a  BioCASE tool for mapping the
data is used to provide the data for GBIF.
BioCASE tool for mapping the
data is used to provide the data for GBIF.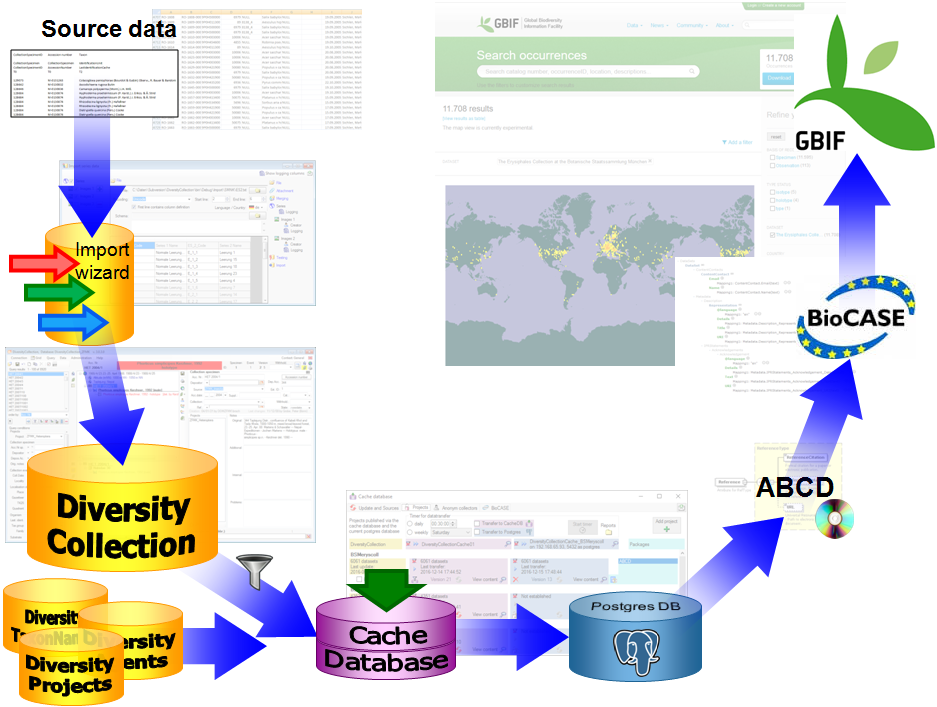
 or export
or export 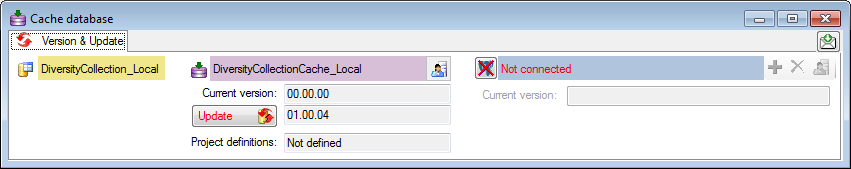
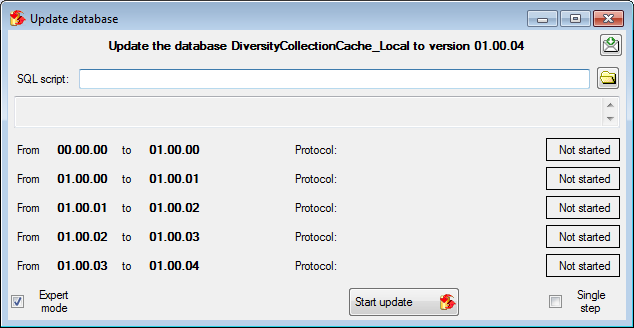

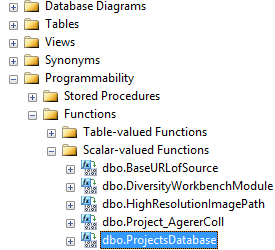

 button to open a window
as shown below. To administrate the access for other logins, you have to
be a System administator. For further details please see the chapter
about the
button to open a window
as shown below. To administrate the access for other logins, you have to
be a System administator. For further details please see the chapter
about the 
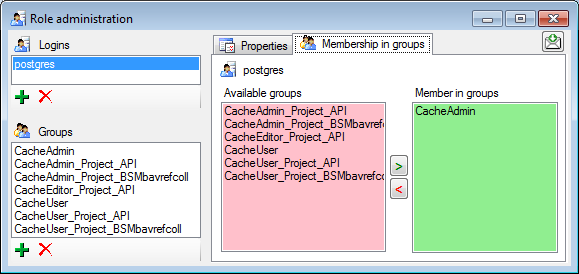
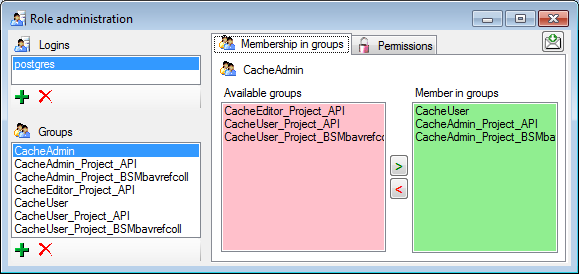
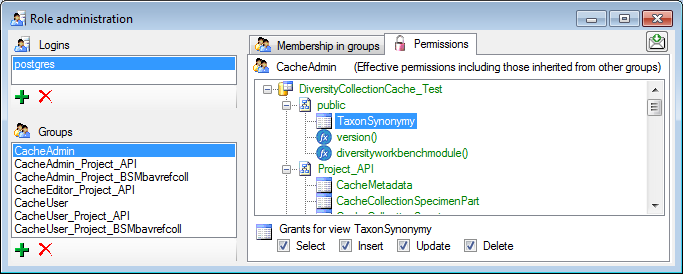
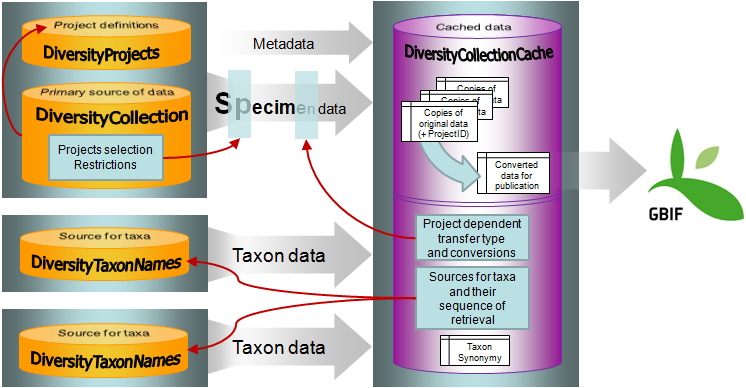
 DiversityTaxonNames). To provide this information add all
sources used in your collection data and transfer the corresponding data
into the cache database. The data in the taxonomic sources are organized
by projects, thus, you need to provide the sequence of the projects that
should be imported into the cache database for every source. A name will
be imported only once. This means that the name with synonymy to the
first imported project will be imported, all following data with this
name will be ignored.
DiversityTaxonNames). To provide this information add all
sources used in your collection data and transfer the corresponding data
into the cache database. The data in the taxonomic sources are organized
by projects, thus, you need to provide the sequence of the projects that
should be imported into the cache database for every source. A name will
be imported only once. This means that the name with synonymy to the
first imported project will be imported, all following data with this
name will be ignored.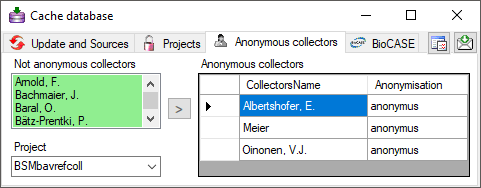
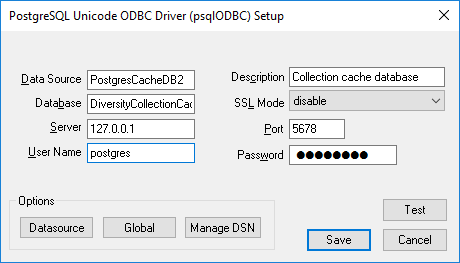
 (see below).
(see below). 
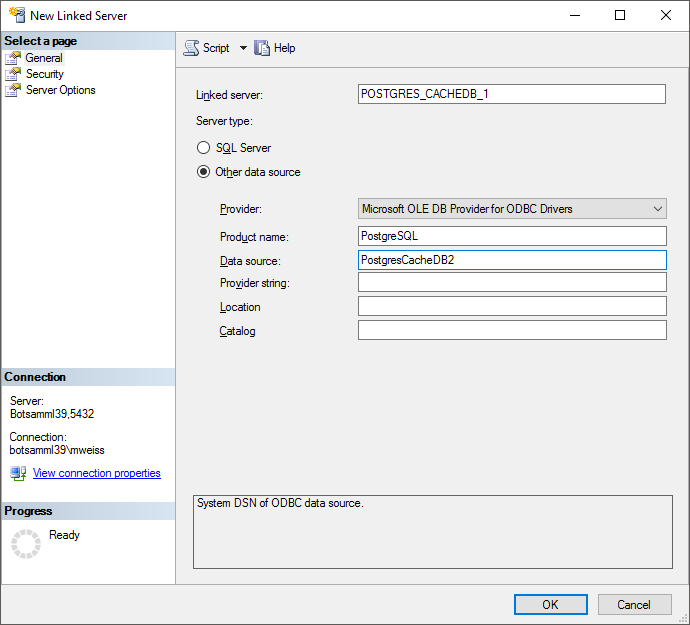
 button.
button.
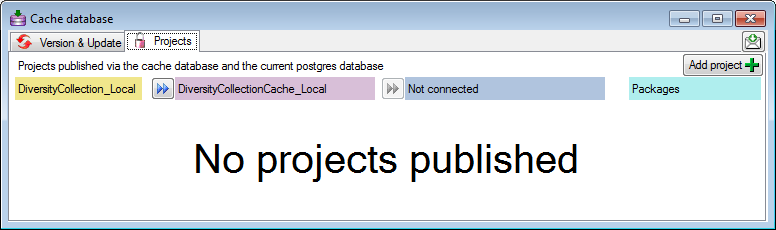

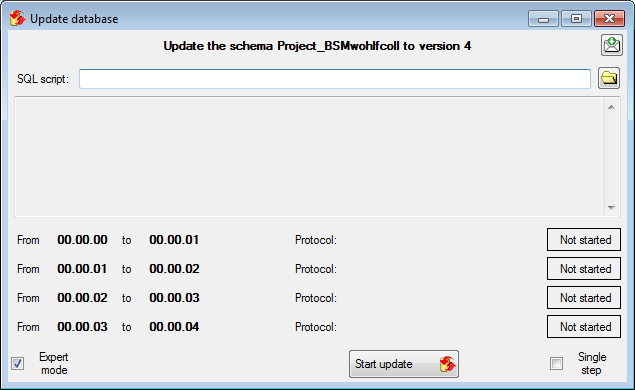
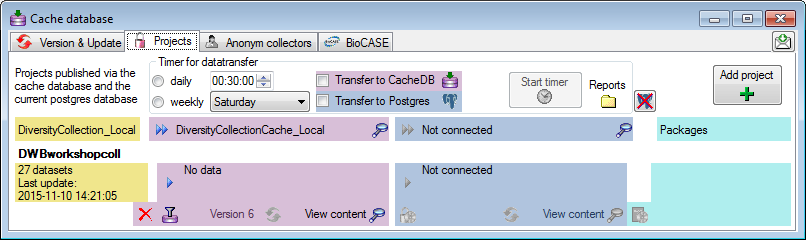
 button and choose the ranges of the
data that should be transferred (see below).
button and choose the ranges of the
data that should be transferred (see below).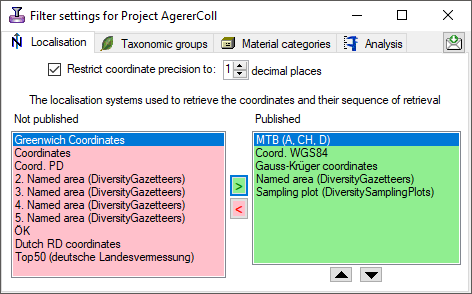
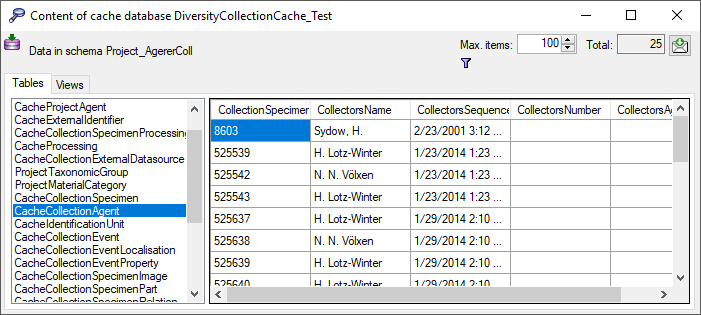
 to the Postgres database and click on the
to the Postgres database and click on the  button to establish
the project and run necessary updates
button to establish
the project and run necessary updates 
 button.
button.

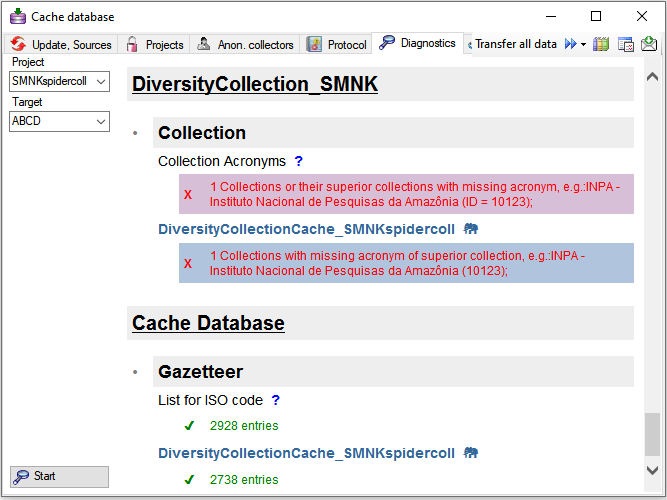
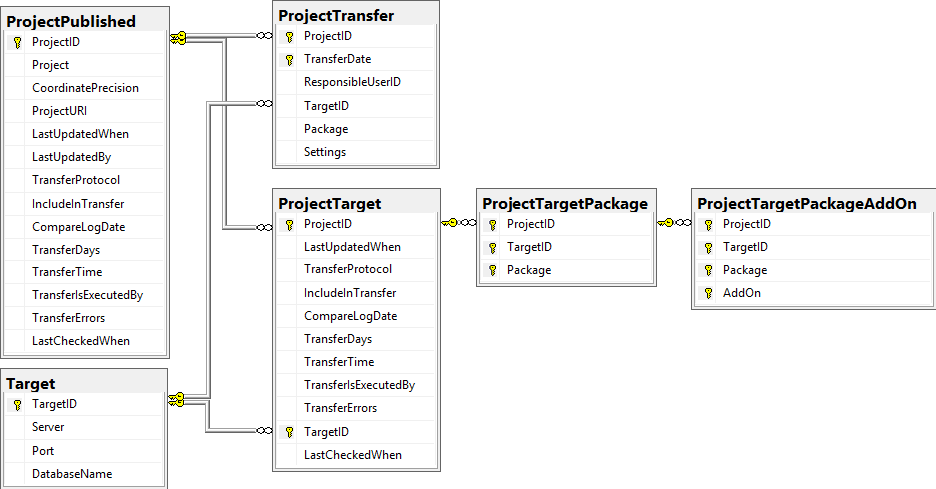


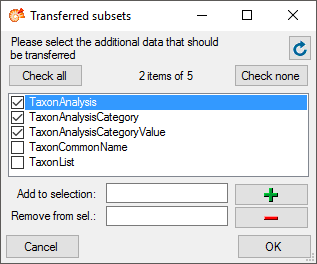
 button will transfer the data of all
sources in the list into the cache database (see below).
button will transfer the data of all
sources in the list into the cache database (see below).
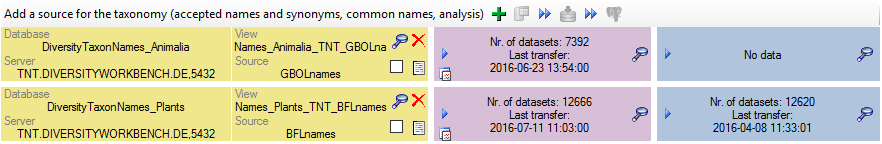

 button (Source database to cache database)
resp.
button (Source database to cache database)
resp.  button (Cache database to Postgres
database). To inspect the data from the views, click on the
button (Cache database to Postgres
database). To inspect the data from the views, click on the
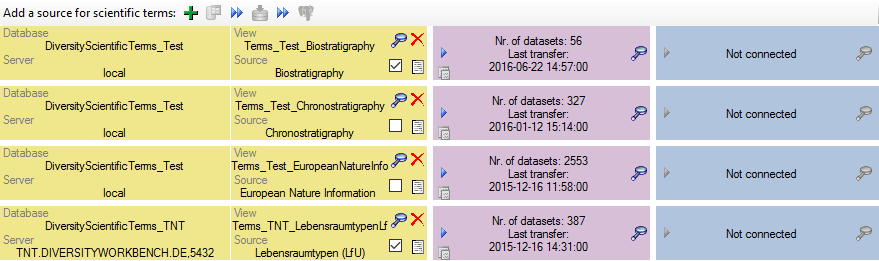
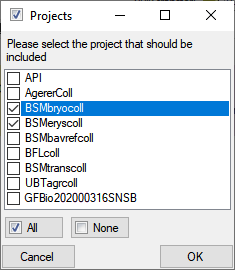

 : Transfer data
of the data
: Transfer data
of the data  . If you transferred only a part
of the data this will be indicated by a thin red border for the current
session
. If you transferred only a part
of the data this will be indicated by a thin red border for the current
session  . The context menu of
the
. The context menu of
the 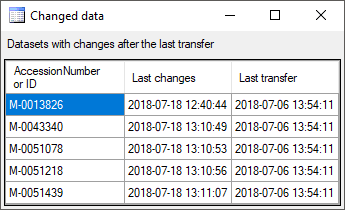

 and the execution will stop in case of an
error. Click on the Start transfer
and the execution will stop in case of an
error. Click on the Start transfer 
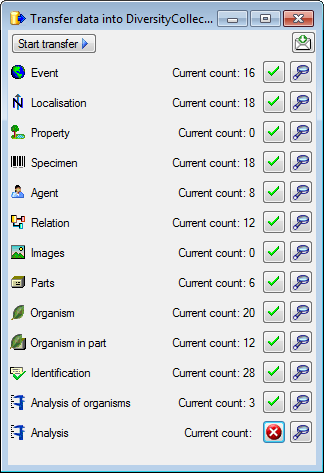
 an error by
an error by
 . The reason for the failure is
shown if you click on the
. The reason for the failure is
shown if you click on the 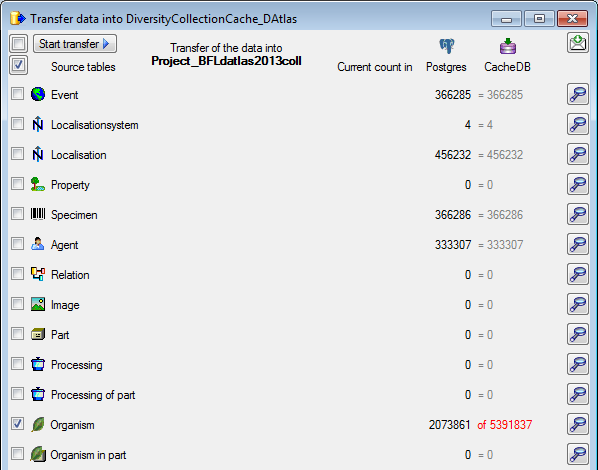

 button in the Timer area to open this directory. To inspect data in the
default schemas (dbo for SQL-Server
button in the Timer area to open this directory. To inspect data in the
default schemas (dbo for SQL-Server  and
public for Postgres
and
public for Postgres 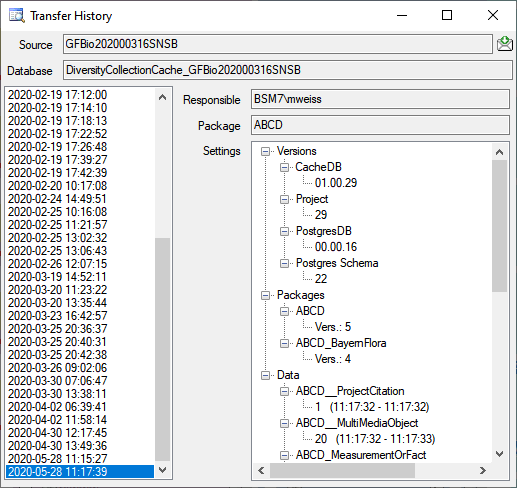
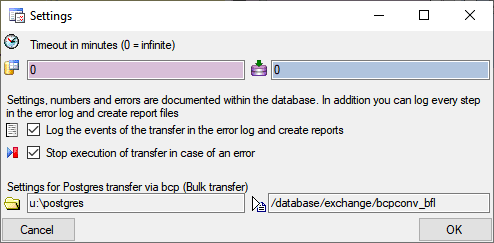


 button will show the protocol in
the default editor of your computer. The
button will show the protocol in
the default editor of your computer. The 

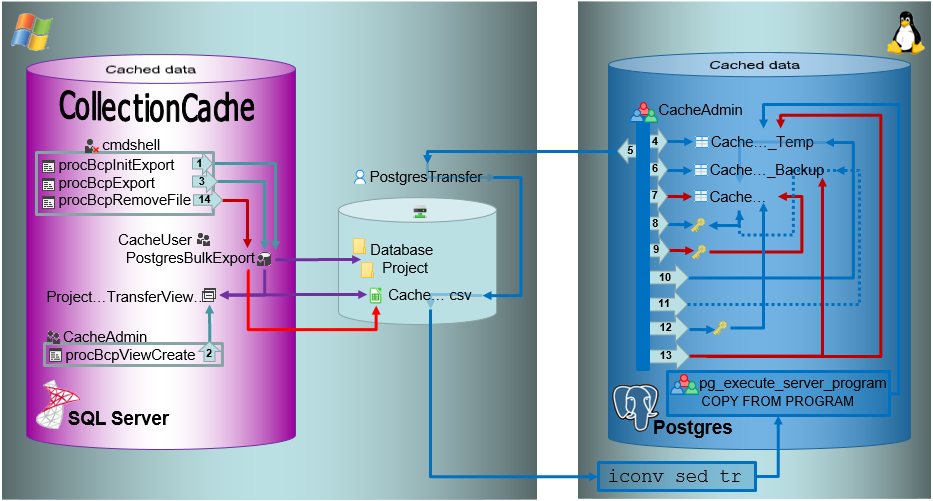

 Computer Management -
Computer Management -
 System-Tools -
System-Tools -
 Local Users and Groups
Local Users and Groups
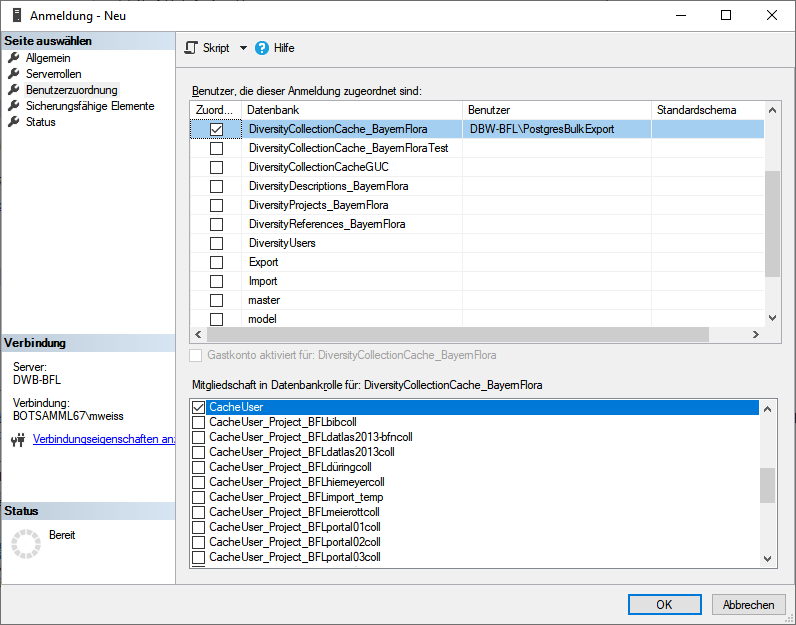

 of the postgres server and the
bashfile
of the postgres server and the
bashfile  executing the transfer must be
in the role
executing the transfer must be
in the role  pg_execute_server_program.
pg_execute_server_program.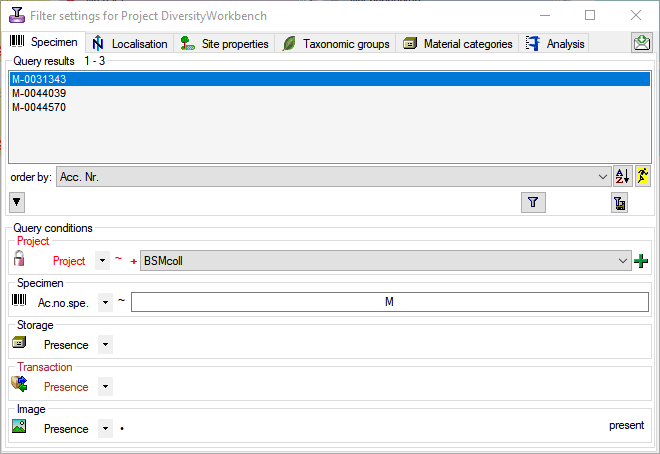
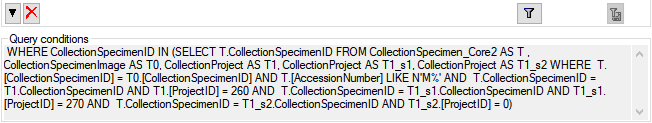

 schemata, with
schemata, with  functions for marking the database as a module of the Diversity
functions for marking the database as a module of the Diversity  TaxonSynonymy where the data derived from DiversityTaxonNames are stored and
TaxonSynonymy where the data derived from DiversityTaxonNames are stored and  views and
views and 

 button. A window will open where you can
enter the new name for the database. Click OK to change the name..
button. A window will open where you can
enter the new name for the database. Click OK to change the name..
 (see below).
(see below).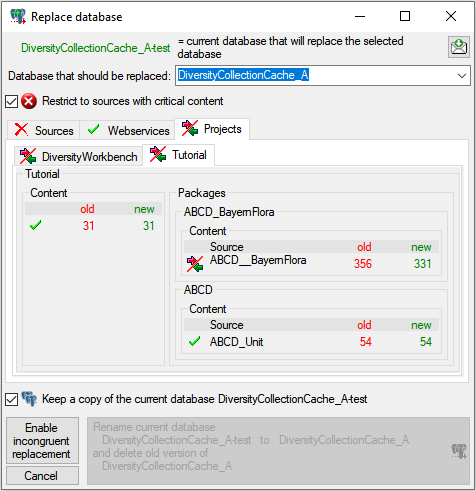

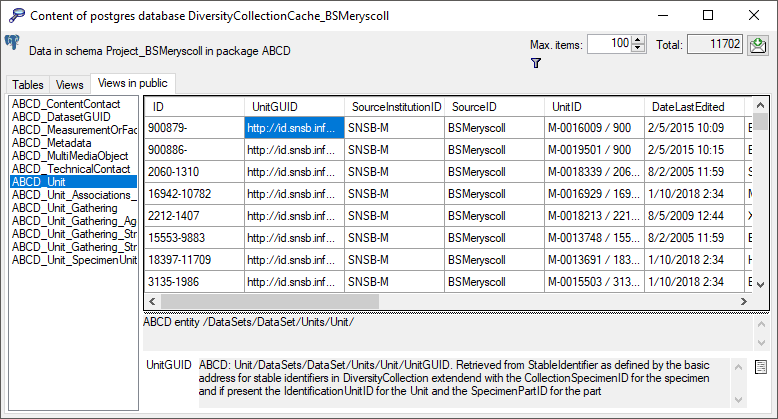
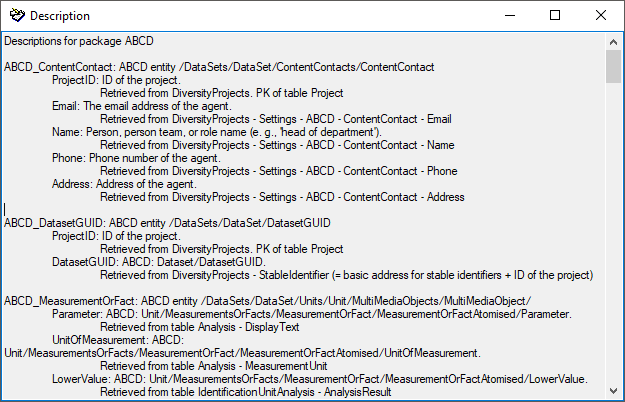
 BioCASE is unable to handle
schemata. Therefore the package ABCD can only be installed for one
project (= schema) in a database. Some packages provide
BioCASE is unable to handle
schemata. Therefore the package ABCD can only be installed for one
project (= schema) in a database. Some packages provide
 SQLite database (creating a SQLite database
containing the tables).
SQLite database (creating a SQLite database
containing the tables).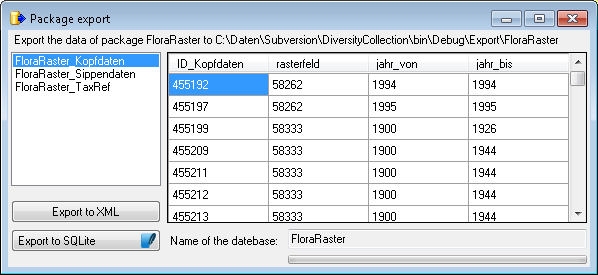
 Transfer data button to update these
views or perform other necessary steps after the data have been
transferred. A window as shown below will open, listing the transfer
steps for the package.
Transfer data button to update these
views or perform other necessary steps after the data have been
transferred. A window as shown below will open, listing the transfer
steps for the package.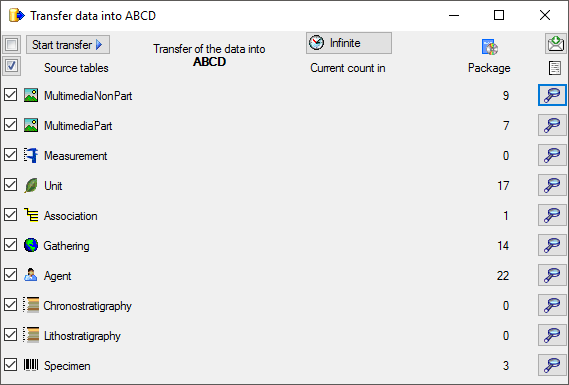
 button as shown below. A window as
shown below will open listing all available add-ons.
button as shown below. A window as
shown below will open listing all available add-ons.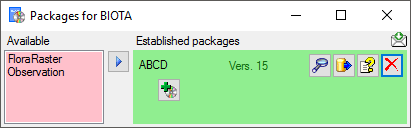
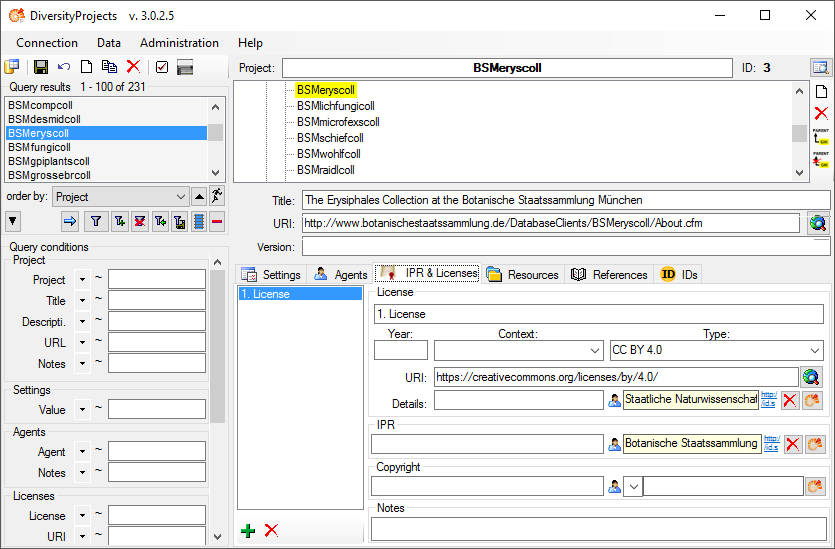
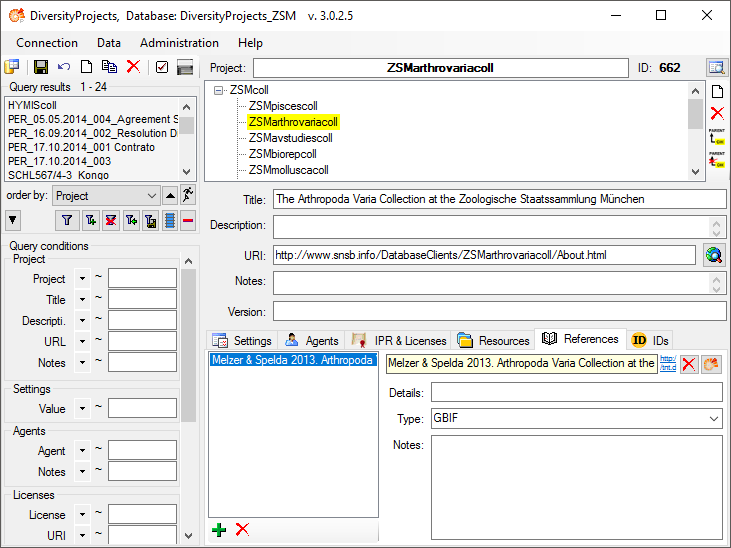
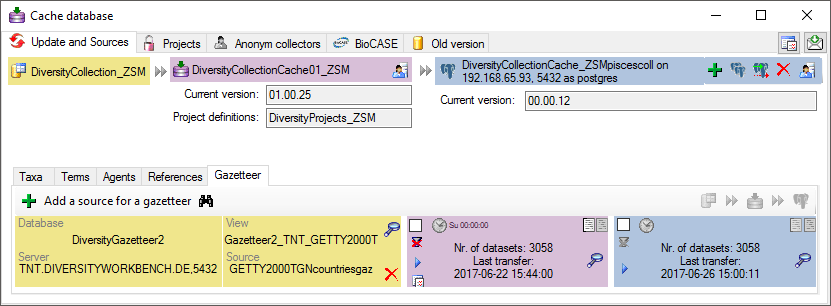
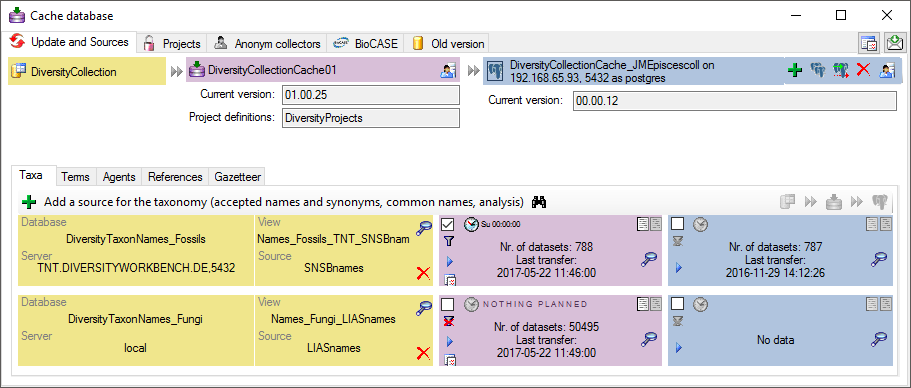

 GFBio to open a window as shown below. Enter
the login data (User + Password) and the Key of the project.
To retrieve the data, click on the Connect
GFBio to open a window as shown below. Enter
the login data (User + Password) and the Key of the project.
To retrieve the data, click on the Connect  button. The software will retrieve the data provided for the project, as
shown below. Information concerning agents will be taken from the
selected DiversityAgents database, and the project selected within this
database. The GFBio portal does not enforce roles for all the agents
entered. If you want to add a role for agents where no role has been given
in the GFBio portal, you may choose a default.
button. The software will retrieve the data provided for the project, as
shown below. Information concerning agents will be taken from the
selected DiversityAgents database, and the project selected within this
database. The GFBio portal does not enforce roles for all the agents
entered. If you want to add a role for agents where no role has been given
in the GFBio portal, you may choose a default.

 DiversityAgents, but are valid for any other module as well.
DiversityAgents, but are valid for any other module as well.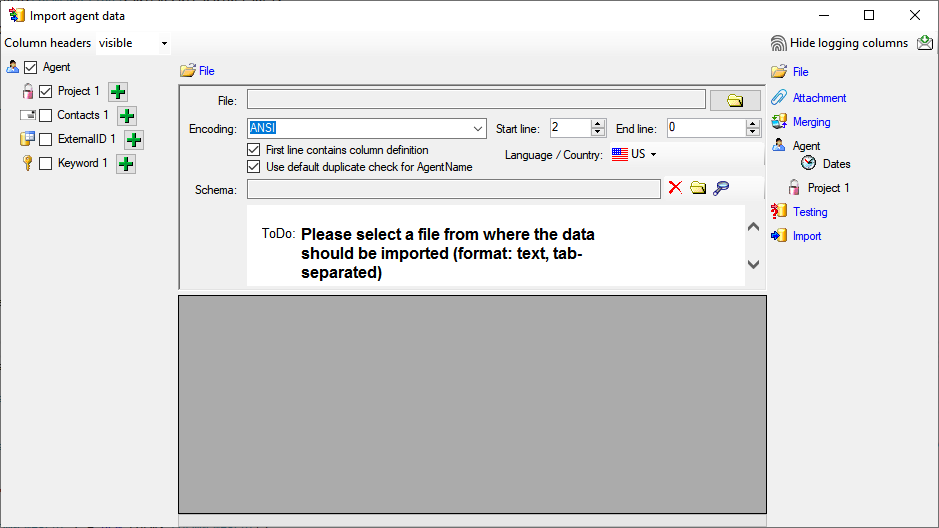
 Record all SQL statements.
Record all SQL statements. 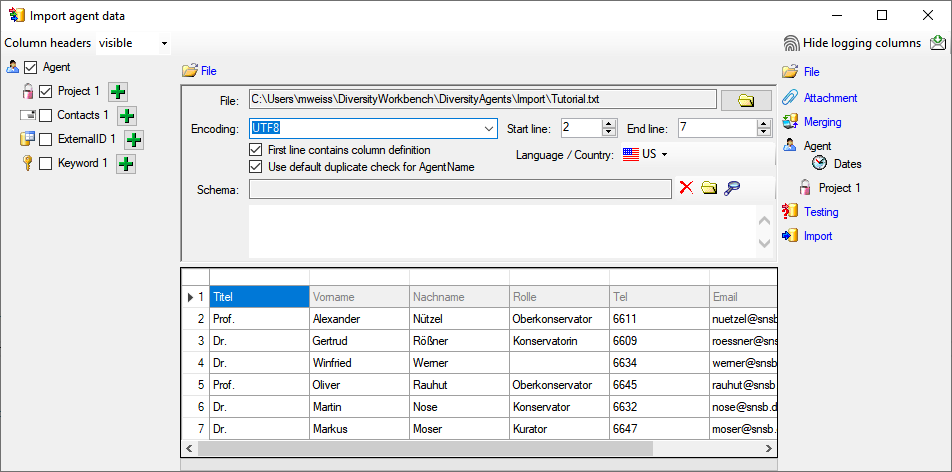
 include logging columns button in the header line. This will include additional substeps for every step containing the
logging columns (see below). If you do not import these data, they will
be automatically filled by default values like the current time and
user.
include logging columns button in the header line. This will include additional substeps for every step containing the
logging columns (see below). If you do not import these data, they will
be automatically filled by default values like the current time and
user.
 Attach
them to data in the database. Select the import step
Attach
them to data in the database. Select the import step

 Merge them with data in the
database. Select the import step
Merge them with data in the
database. Select the import step  Insert,
Insert,  Update and
Update and 
 = If data will be imported depends on the content of decisive
columns, so at least one must be selected.
= If data will be imported depends on the content of decisive
columns, so at least one must be selected. = You have
to select a value from the provided list
= You have
to select a value from the provided list = You have to enter
a value used for all datasets
= You have to enter
a value used for all datasets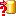 - To test if all requirements for the import are met use the
- To test if all requirements for the import are met use the
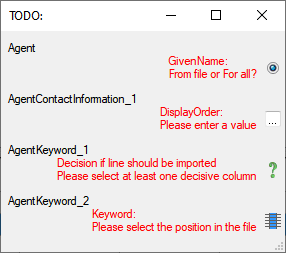
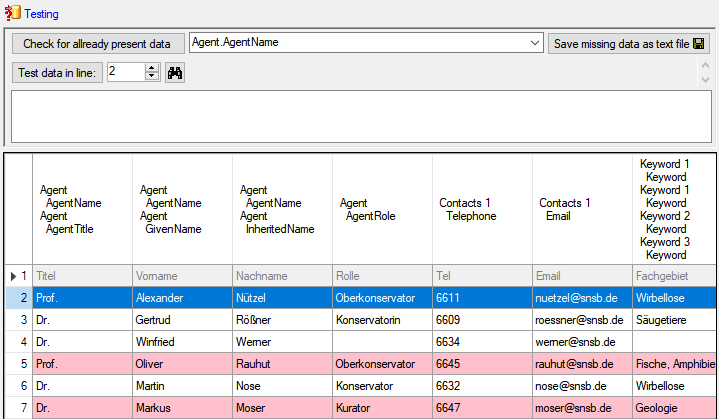

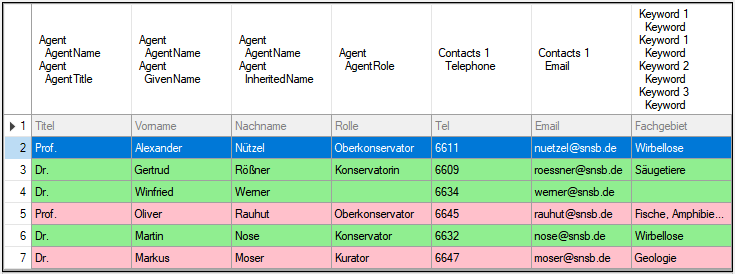
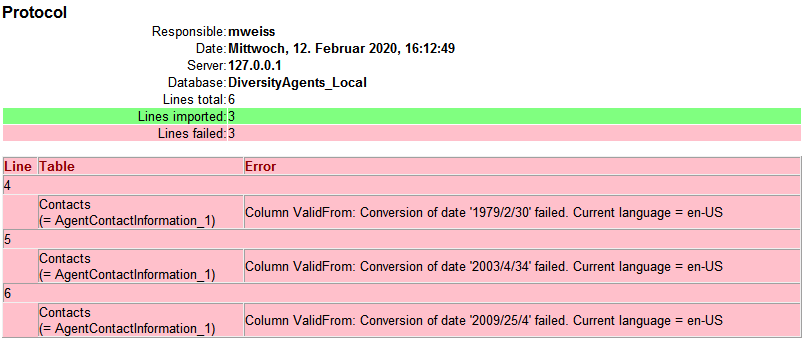
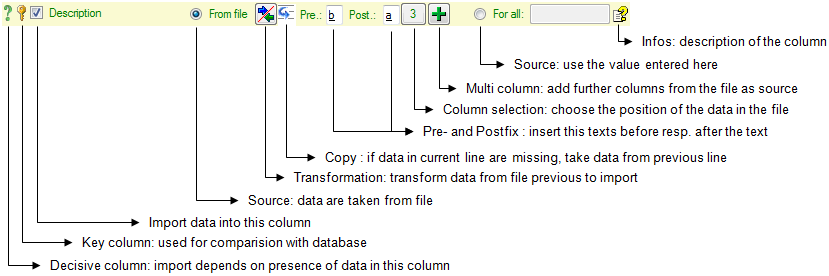

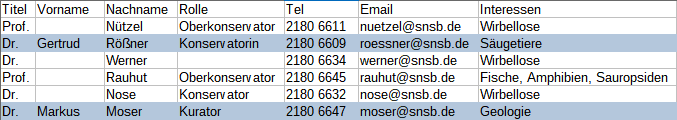
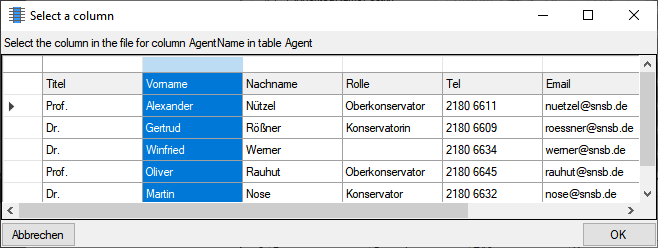
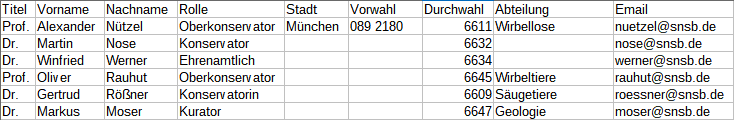
 Copy line option to fill in
missing data as shown below where the blue
values are copied into empty fields during the
import. Click on the
Copy line option to fill in
missing data as shown below where the blue
values are copied into empty fields during the
import. Click on the 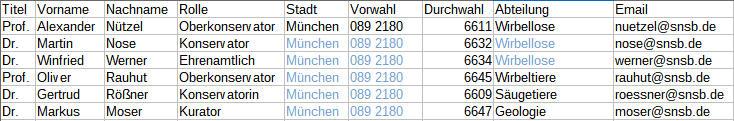
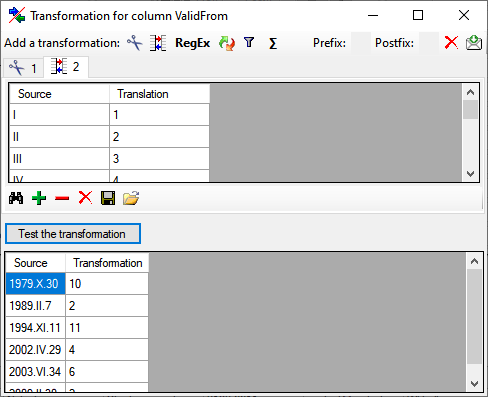
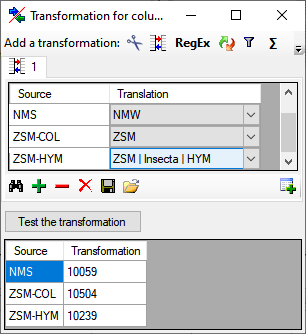
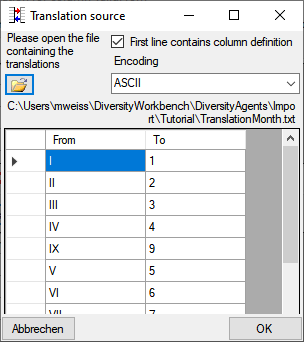
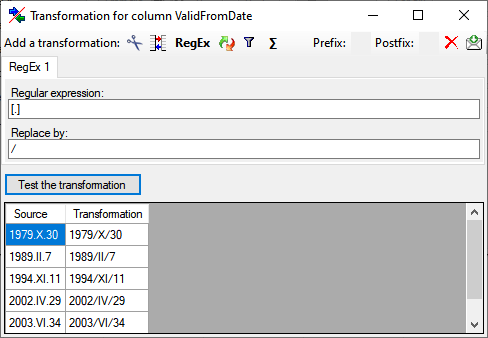
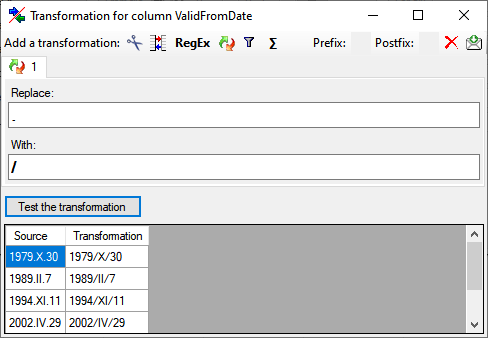
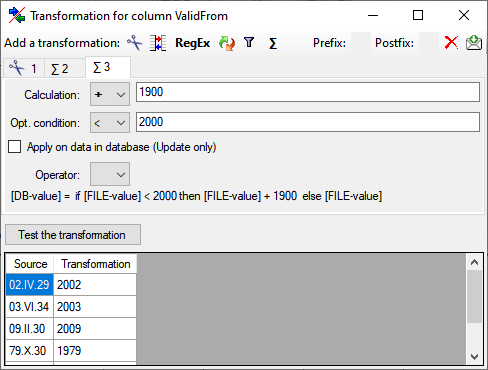
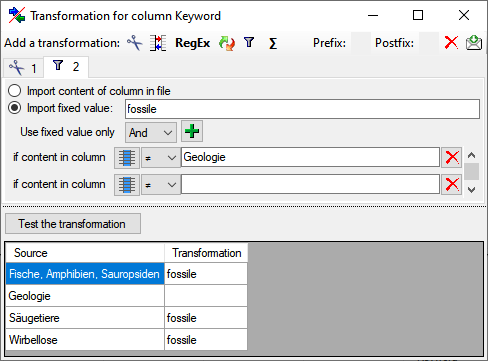
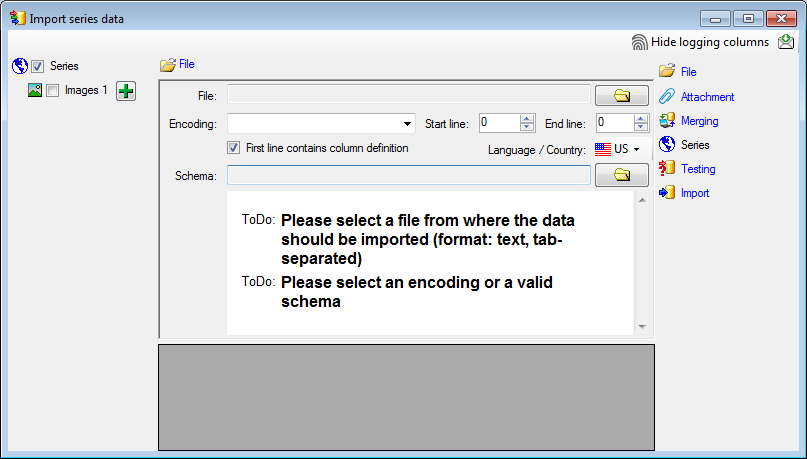
 button in the header line.
This will include a additional substeps for every step containing the
logging columns (see below). If you do not import these data, they will
be automatically filled by default values like the current time and
user.
button in the header line.
This will include a additional substeps for every step containing the
logging columns (see below). If you do not import these data, they will
be automatically filled by default values like the current time and
user.


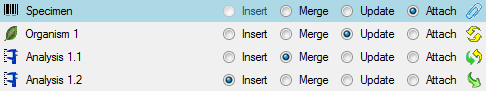
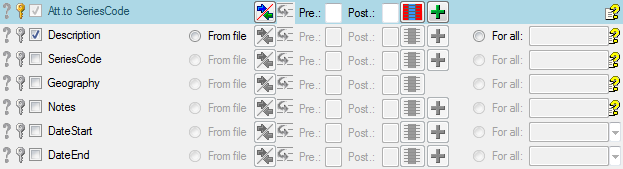
 = You have to enter
a value used for all datasets
= You have to enter
a value used for all datasets Testing step. You can use a certain line in
the file for you test and than click on the Test data in line:
button. If there are still unmet requirements, these will be listed in a
window as shown below.
Testing step. You can use a certain line in
the file for you test and than click on the Test data in line:
button. If there are still unmet requirements, these will be listed in a
window as shown below.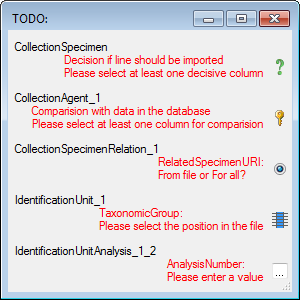
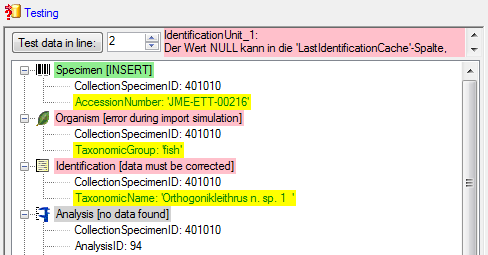
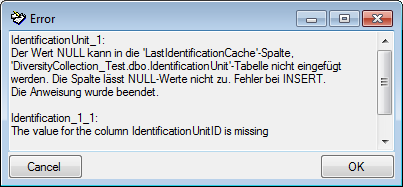
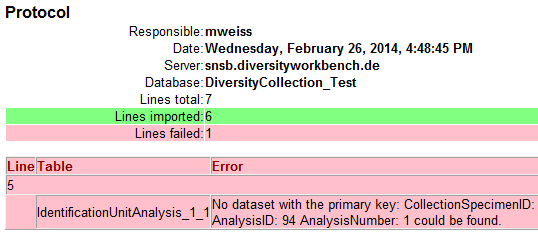
 Replicator or Administrator.
Replicator or Administrator.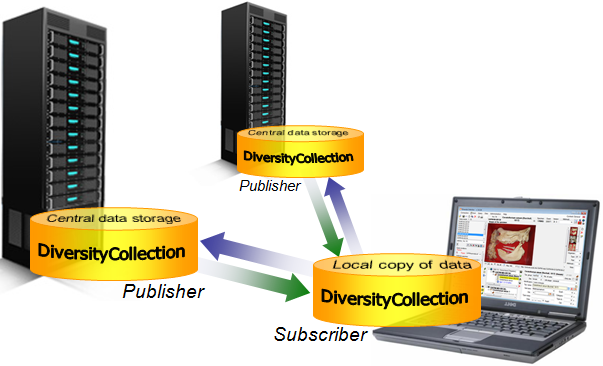
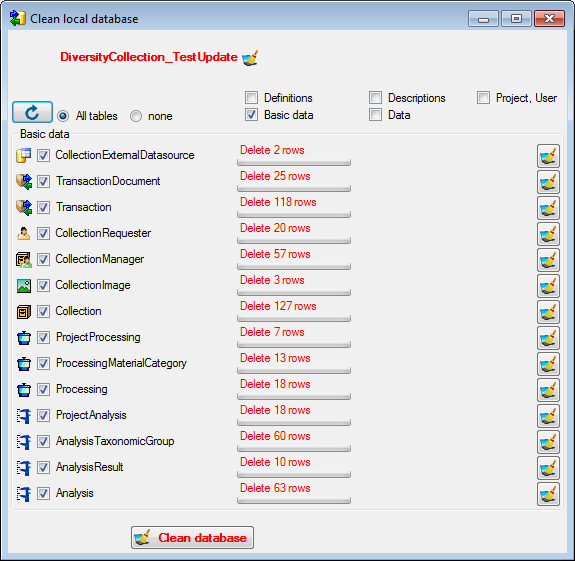
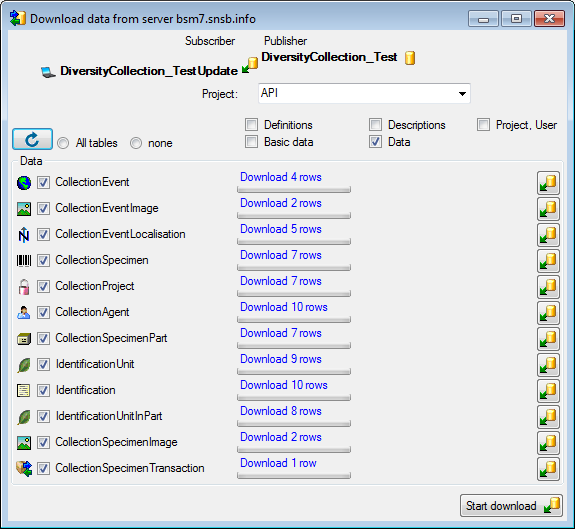
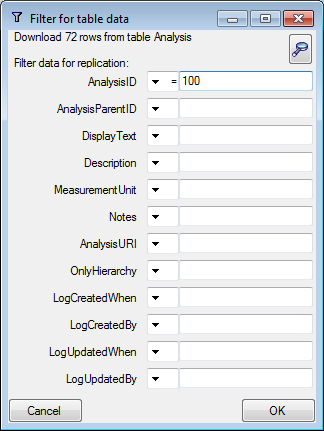
 .
.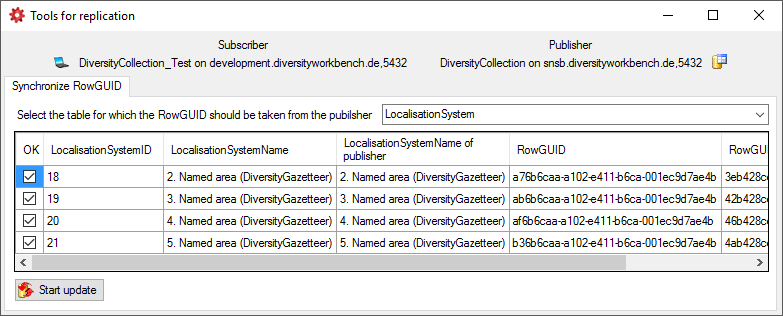


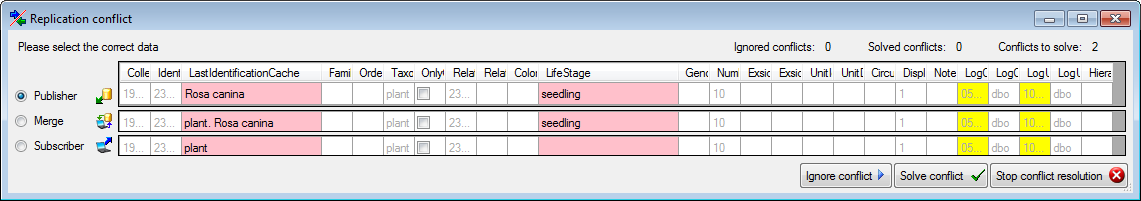
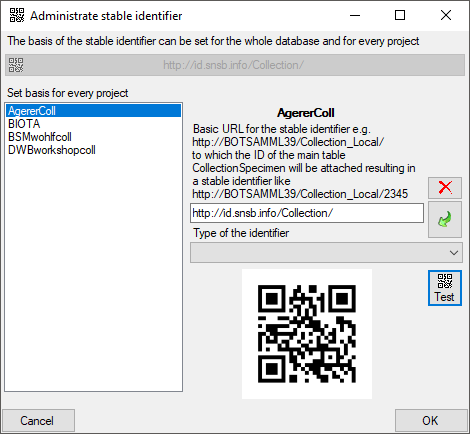
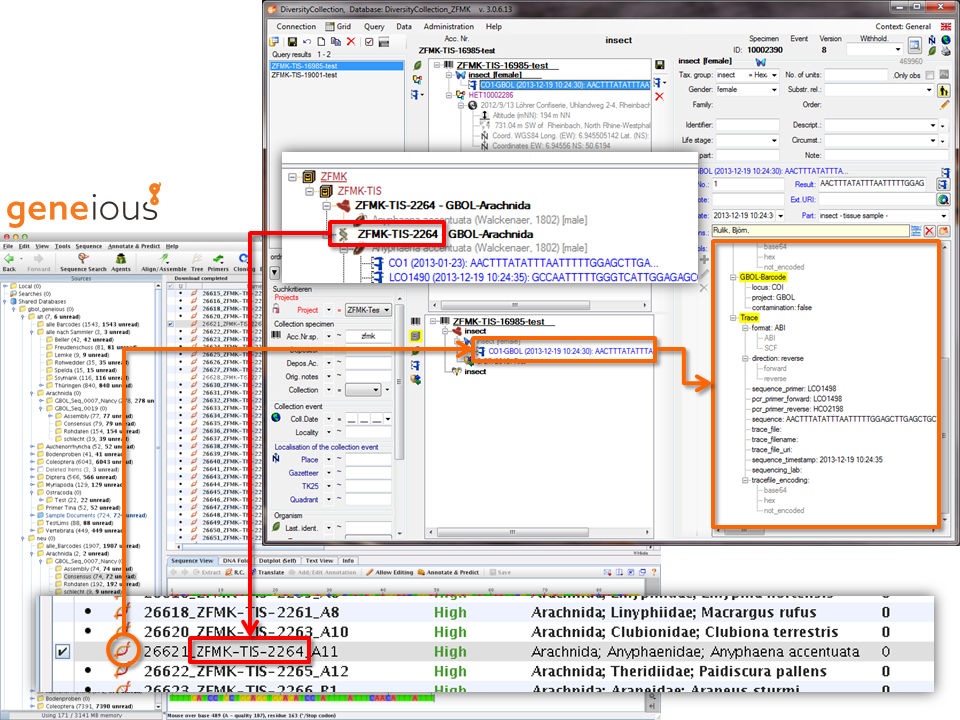
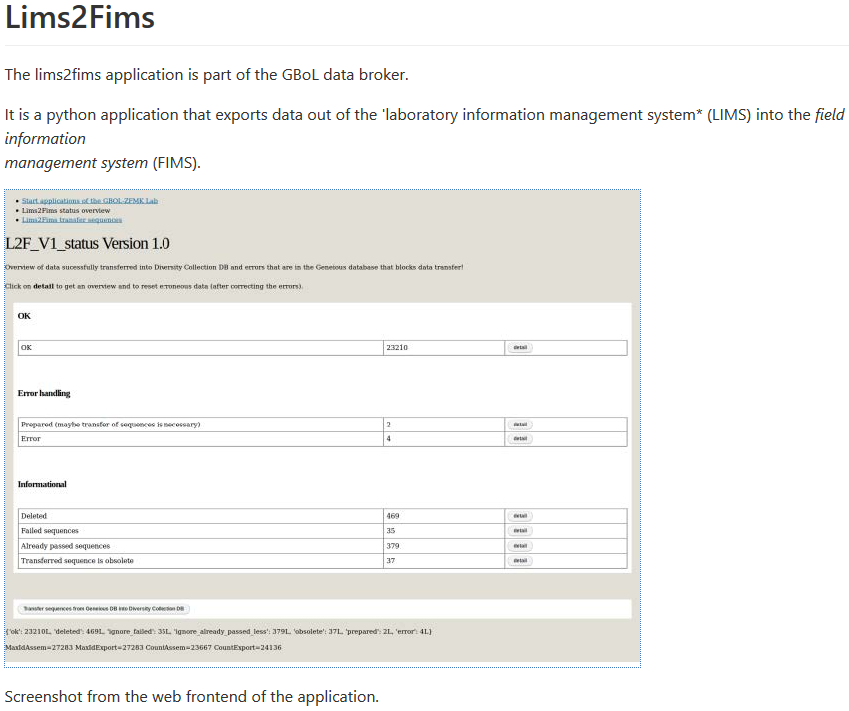





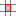

 Box import
Box import  Areas import
Areas import  Taxa import
Taxa import



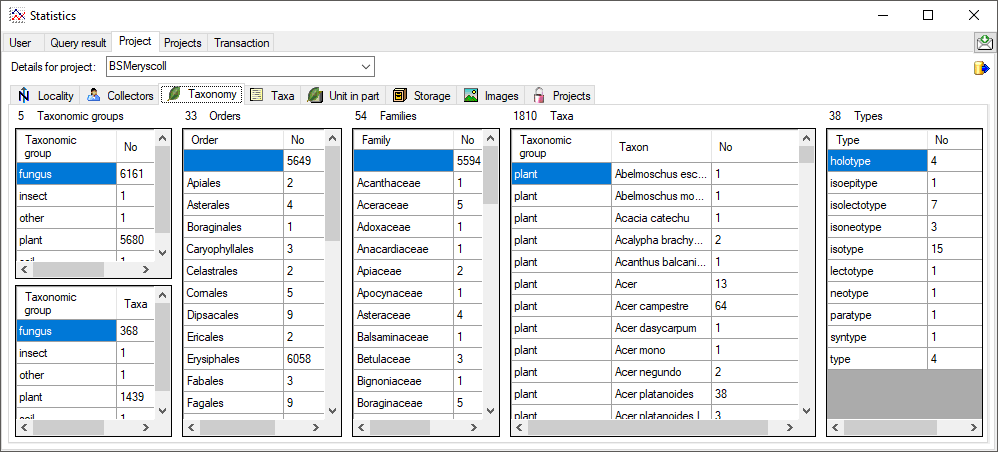
 from the menu. A window will open as
shown below where you may edit the entities defined for the program.
from the menu. A window will open as
shown below where you may edit the entities defined for the program.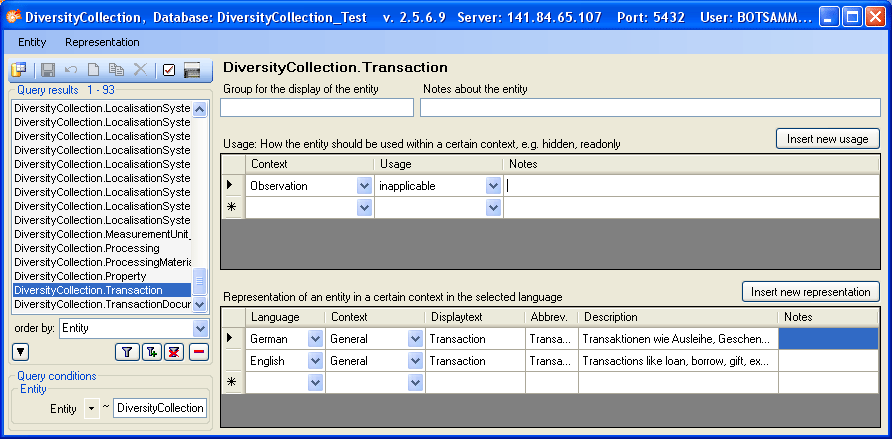
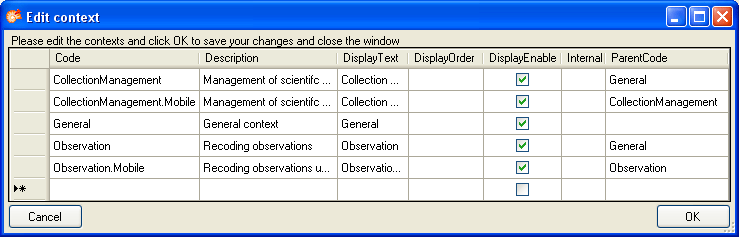
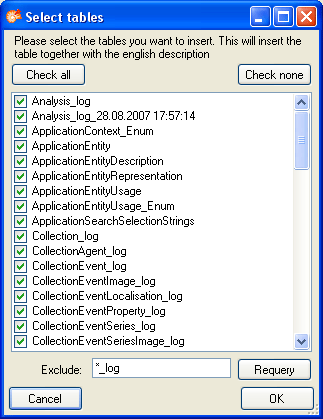


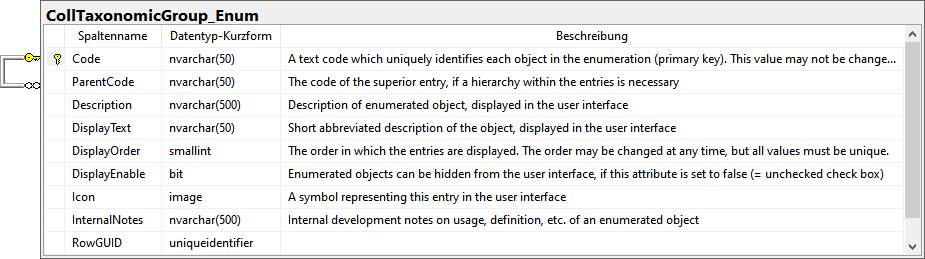
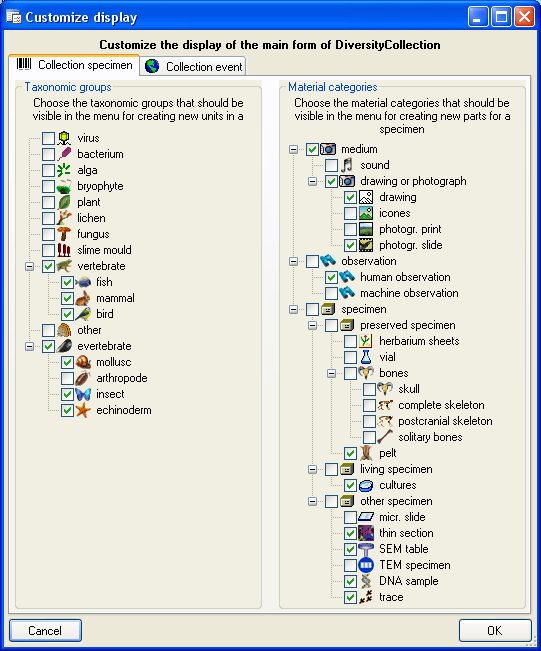
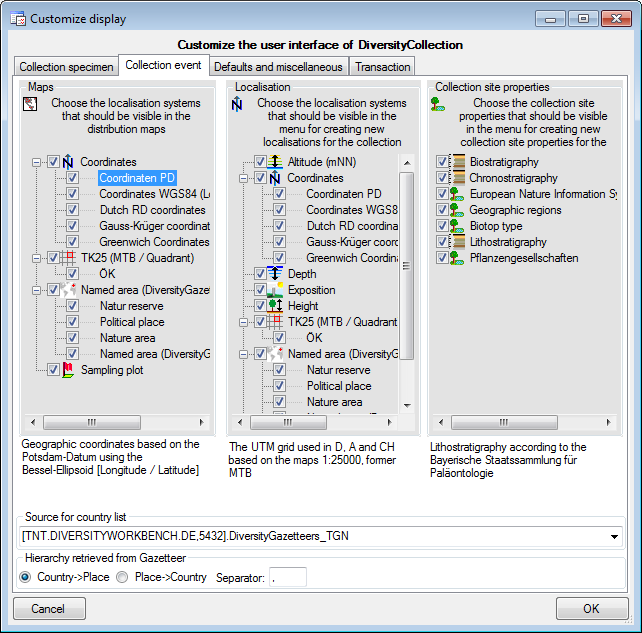
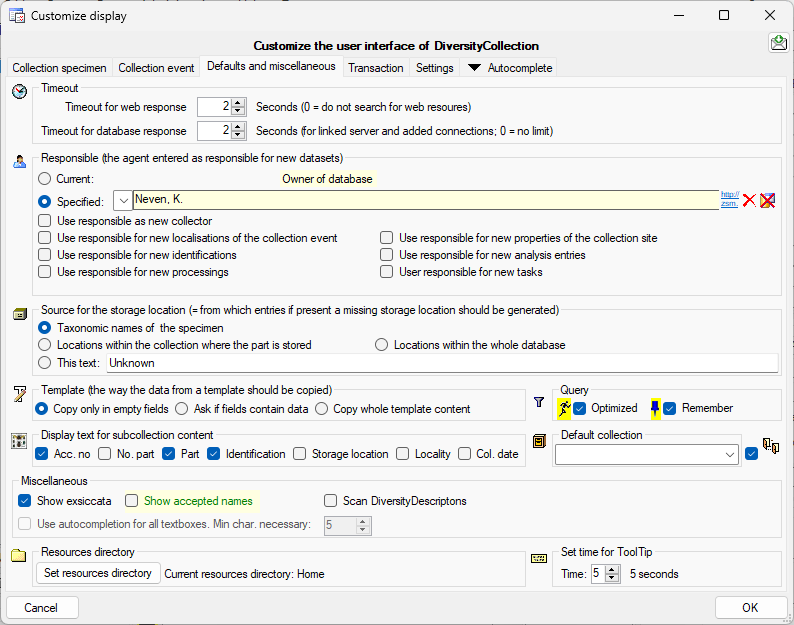
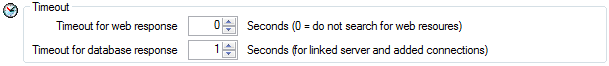

 Use translations: If
the data fields for the
Use translations: If
the data fields for the 
 Load datasources: If
the datasources on
Load datasources: If
the datasources on 
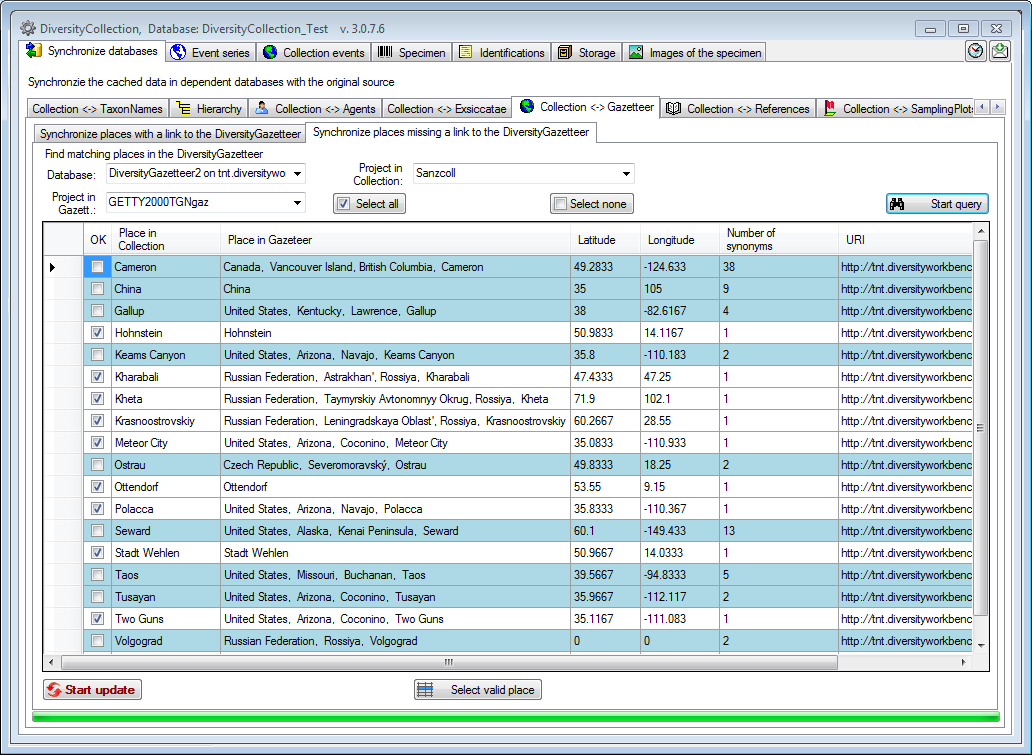
 Select valid place button and select the correct name from the list
as shown below.
Select valid place button and select the correct name from the list
as shown below.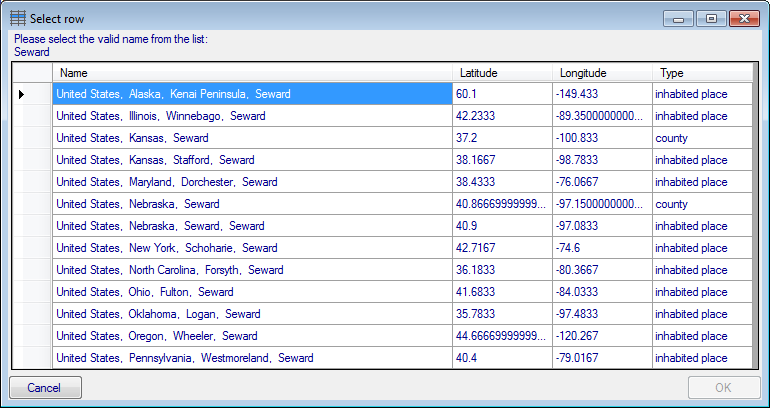
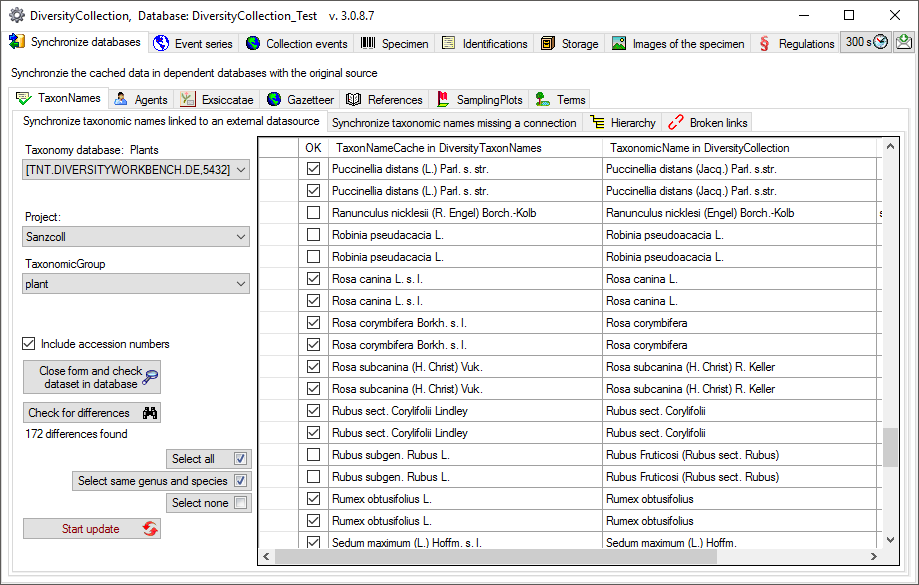
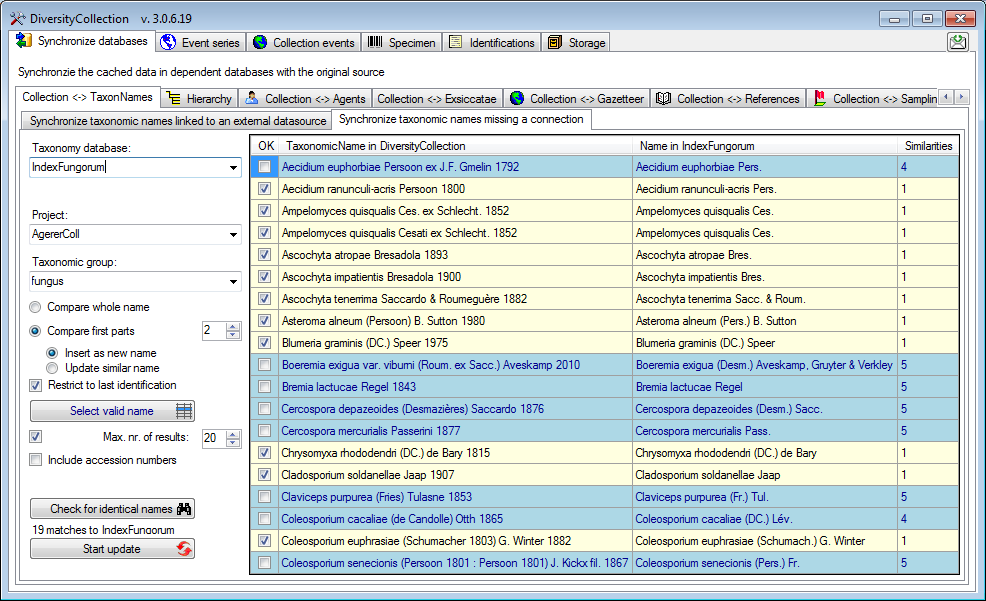
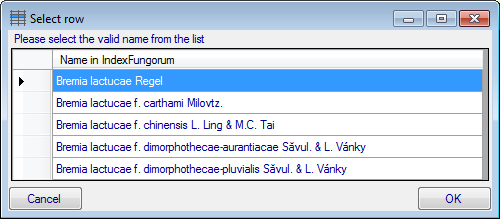
 , e.g. when a webservice changes its address, you can search for these (see below).
Please make sure, that you are
, e.g. when a webservice changes its address, you can search for these (see below).
Please make sure, that you are 
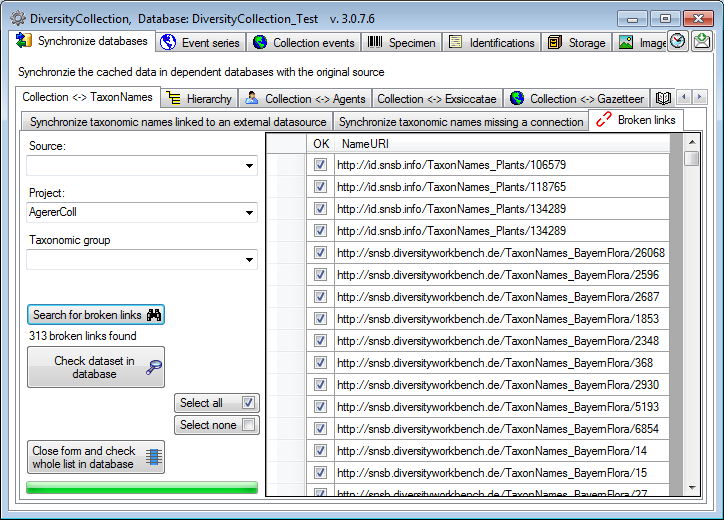
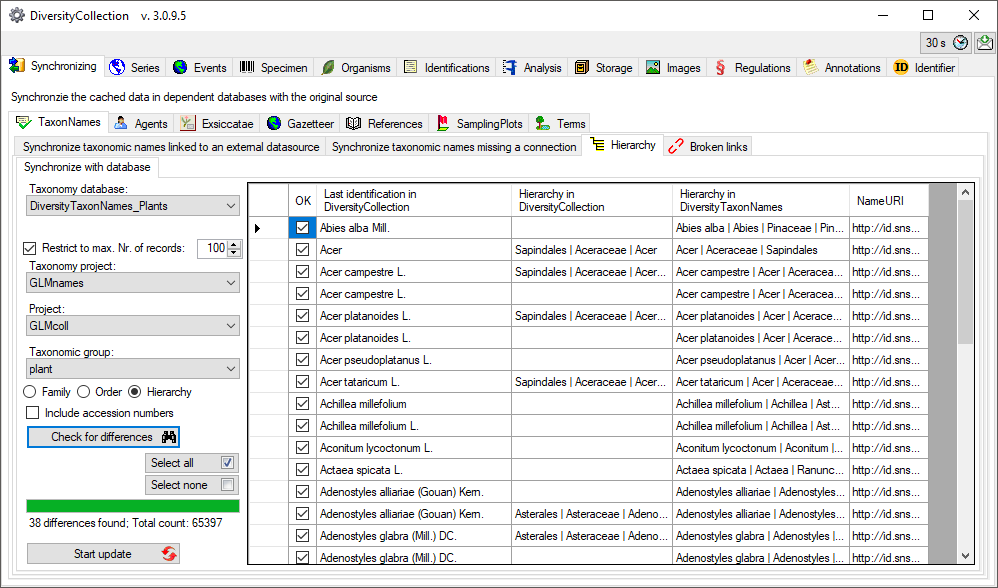

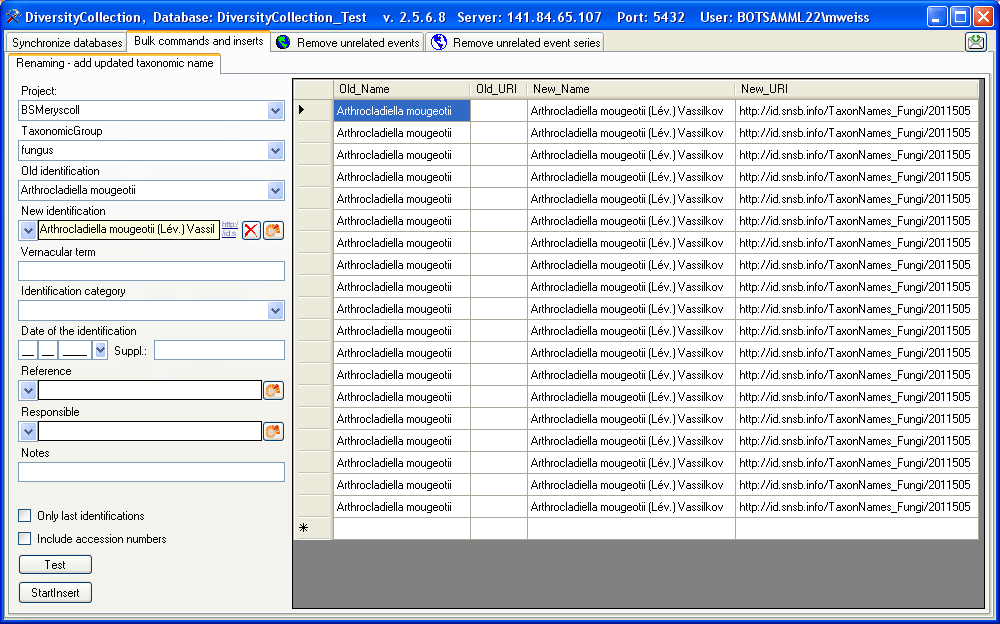
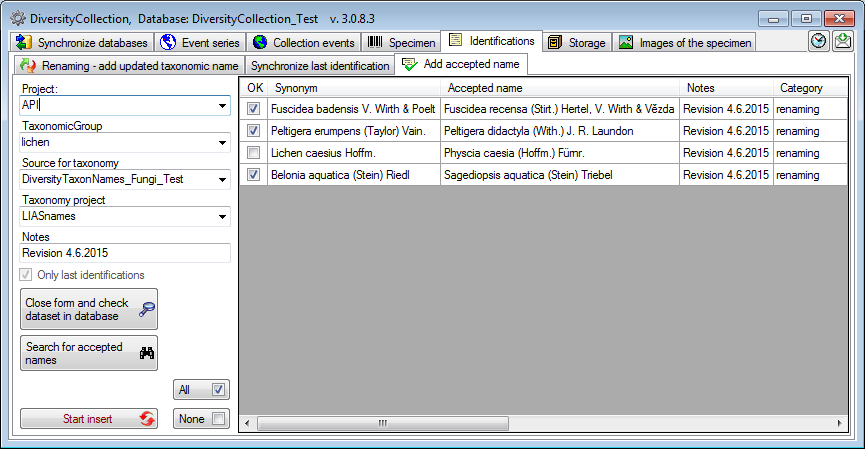
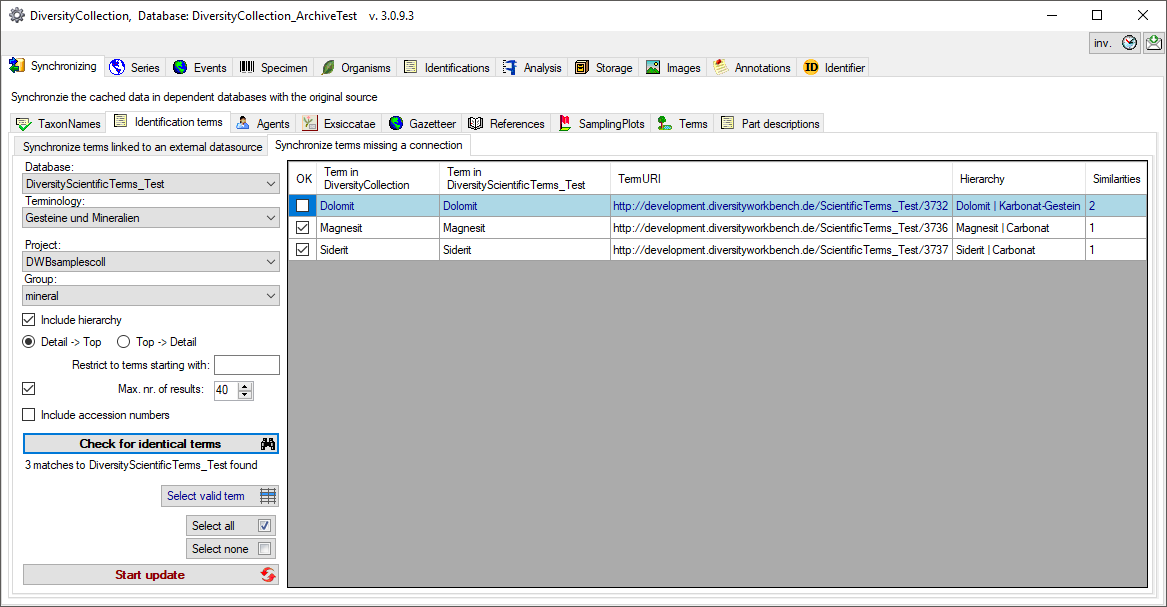
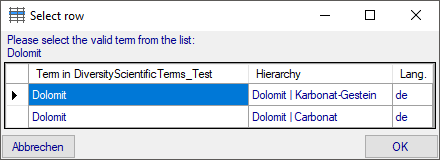
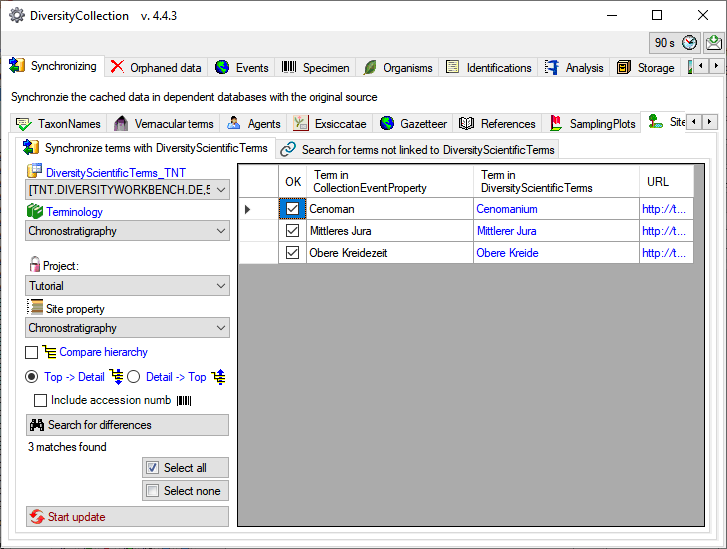
 or
or  . As shown below 2 more columns will be
included, containing the hierarchies in DiversityCollection and
. As shown below 2 more columns will be
included, containing the hierarchies in DiversityCollection and 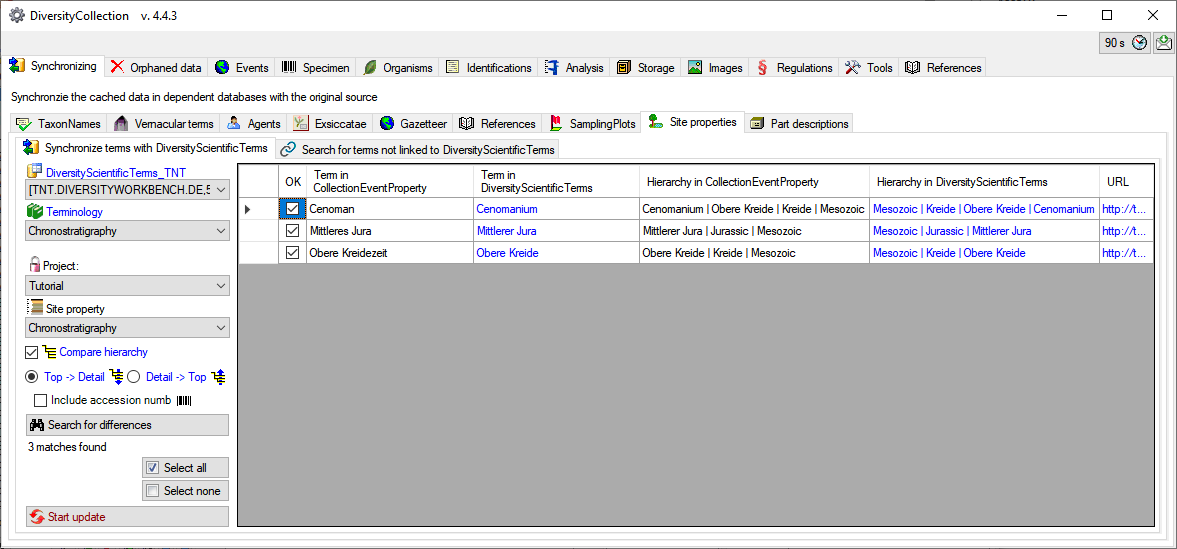
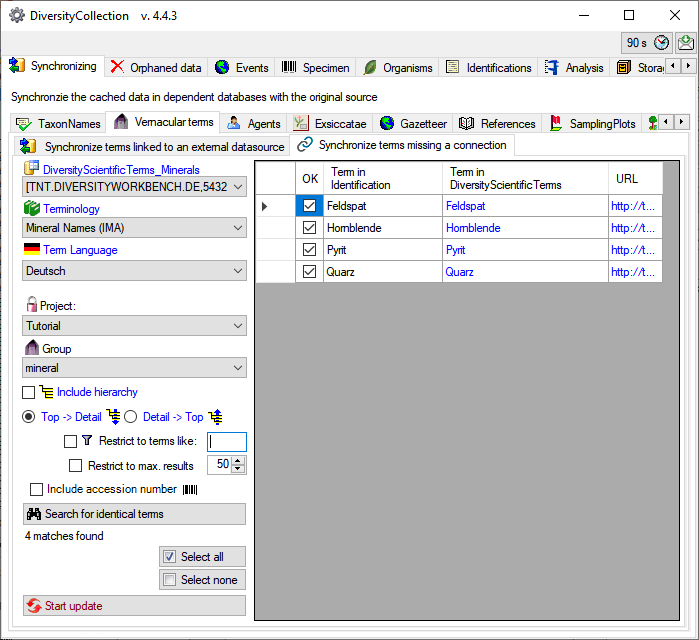
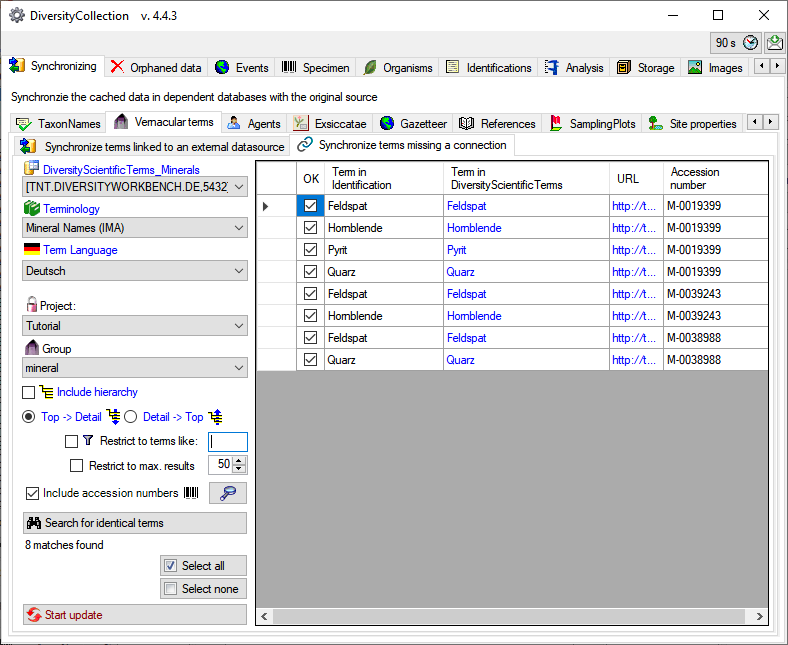
 button. The type name is written
as defined by the default display type in the module DiversityAgents -
for more information please see the documentation on DiversityAgents.
button. The type name is written
as defined by the default display type in the module DiversityAgents -
for more information please see the documentation on DiversityAgents.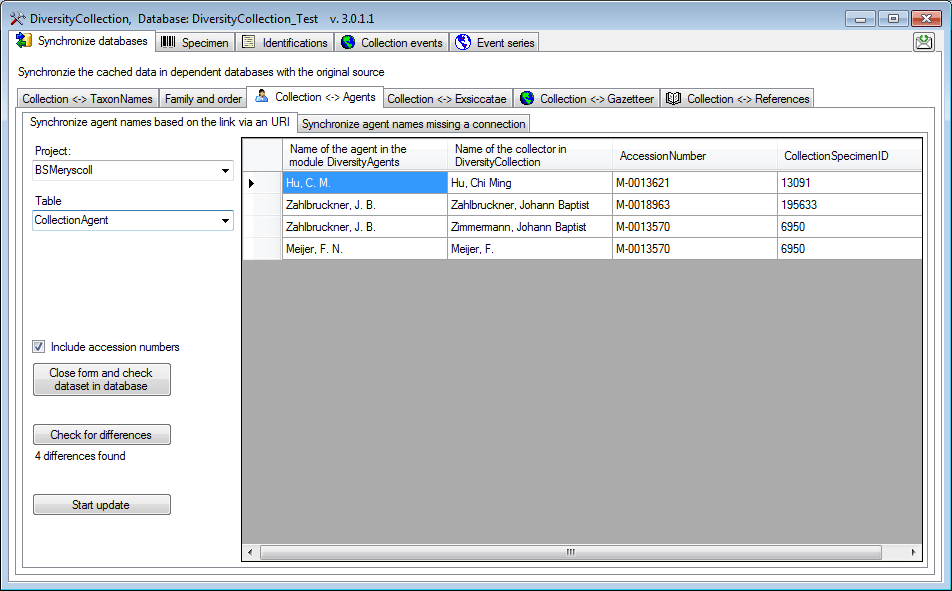
 button. If you wish to have a more
detailed look on one of the data sets in the list, you need to check the
“Include accession number” checkbox before starting the query. A button
button. If you wish to have a more
detailed look on one of the data sets in the list, you need to check the
“Include accession number” checkbox before starting the query. A button
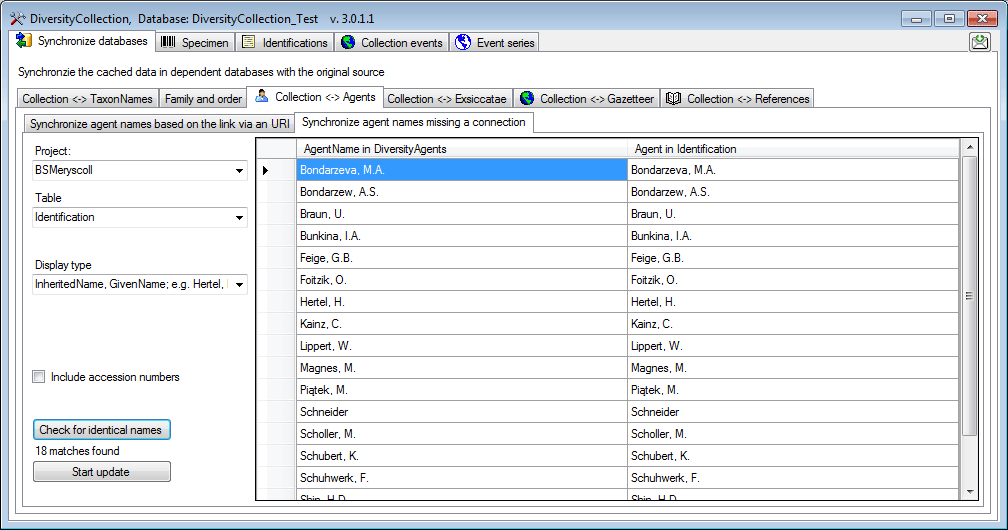
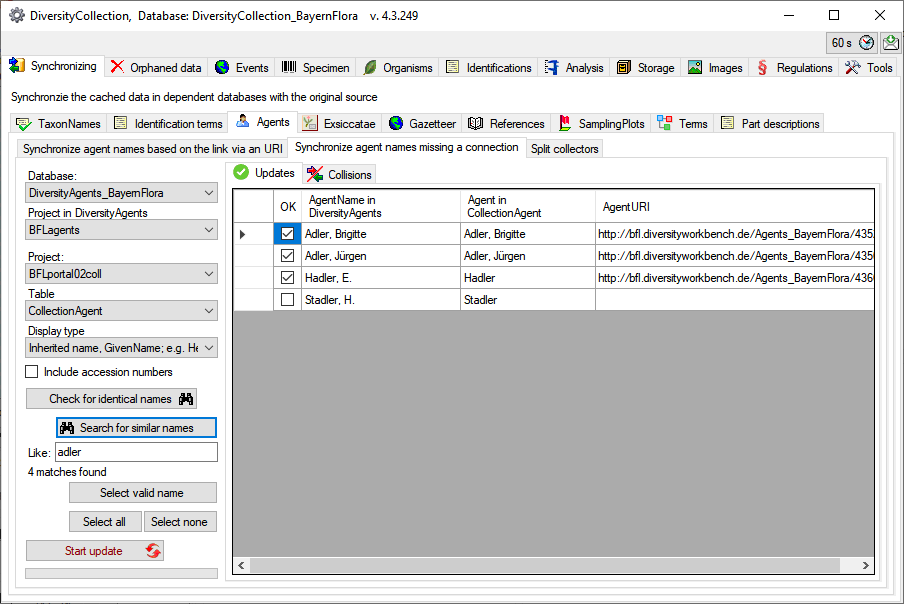
 site property for which the entries should be synchronized.
site property for which the entries should be synchronized.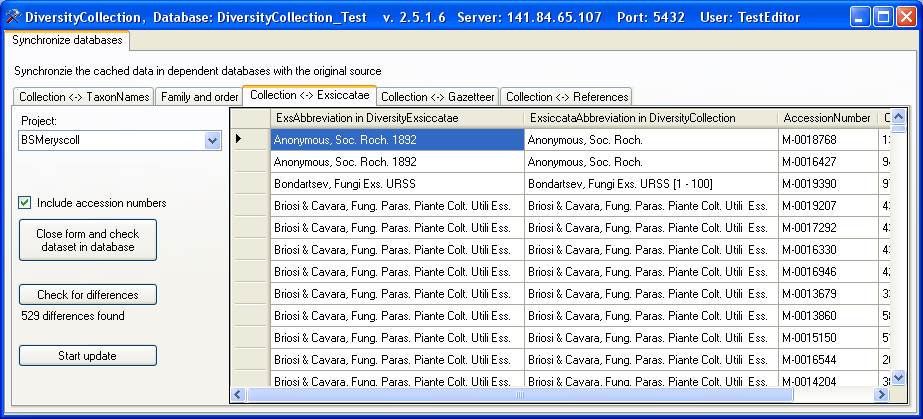
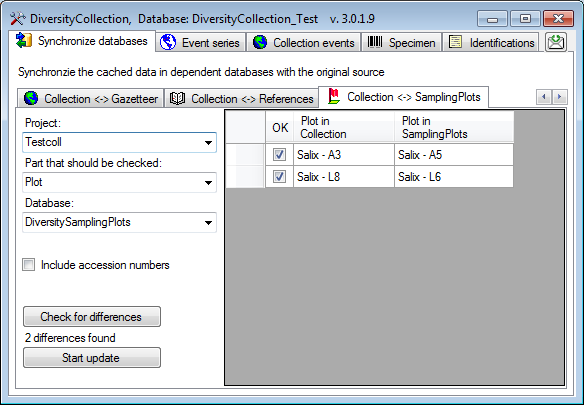
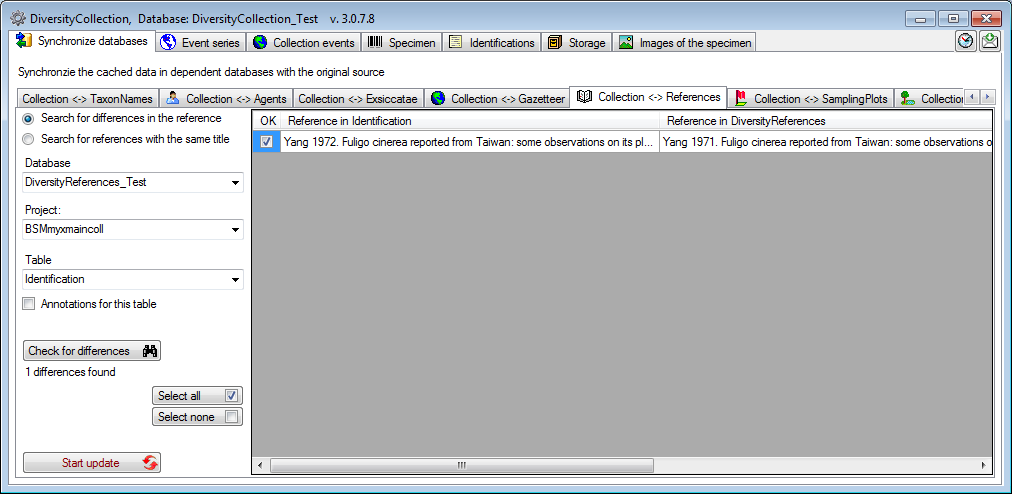
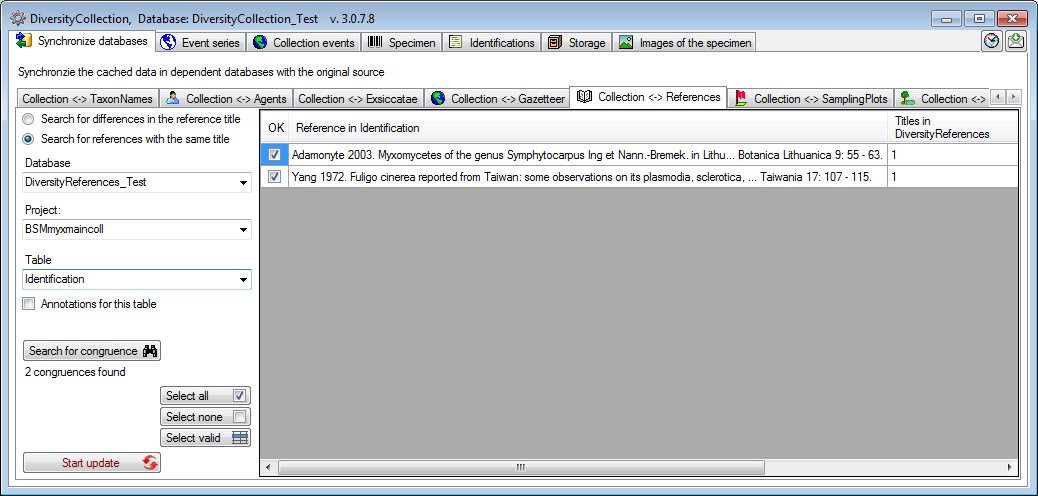
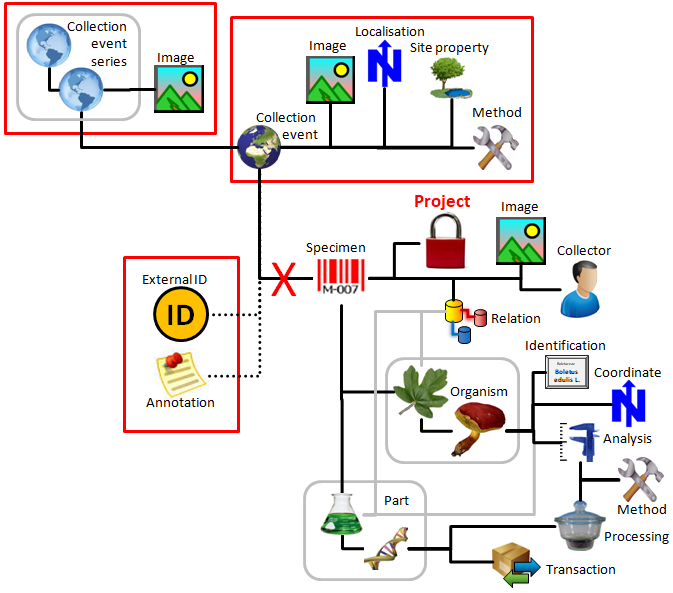
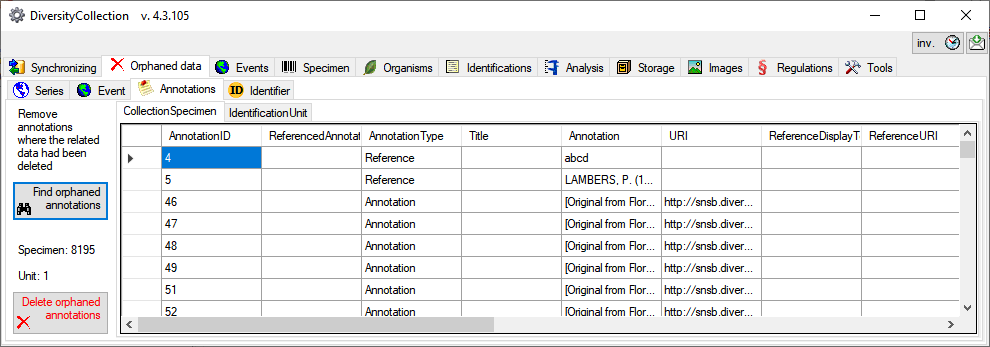
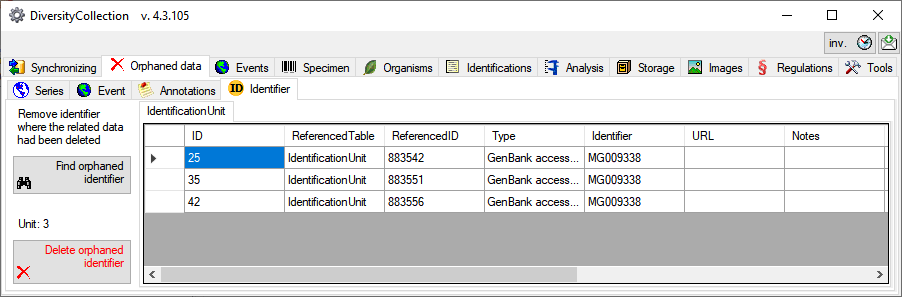
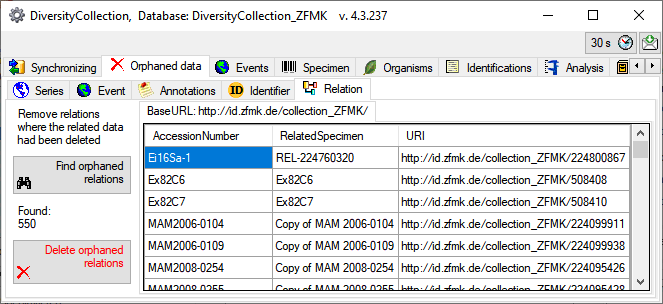
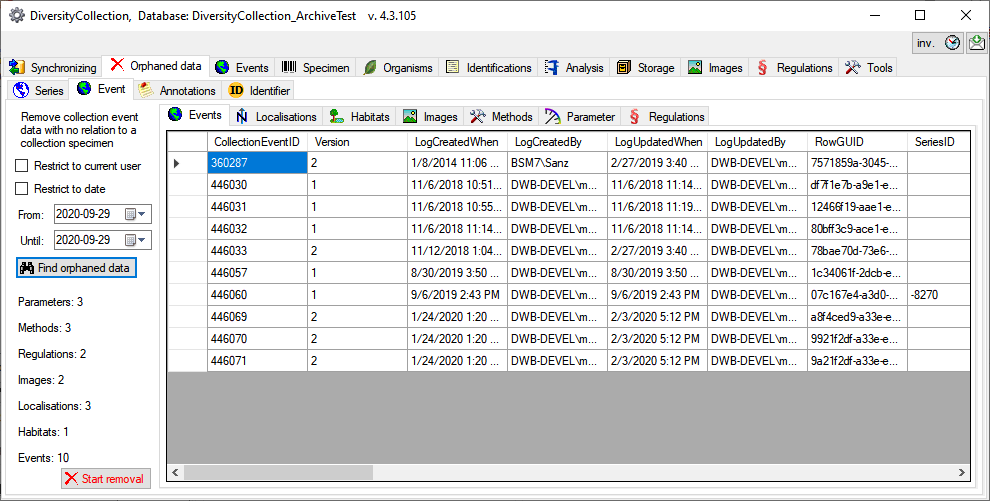
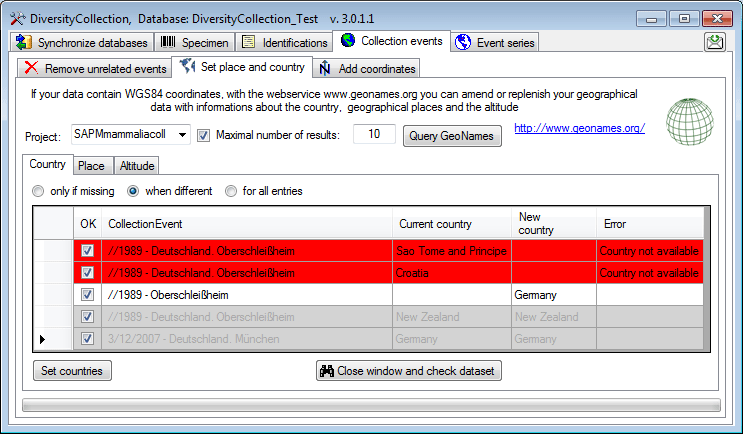
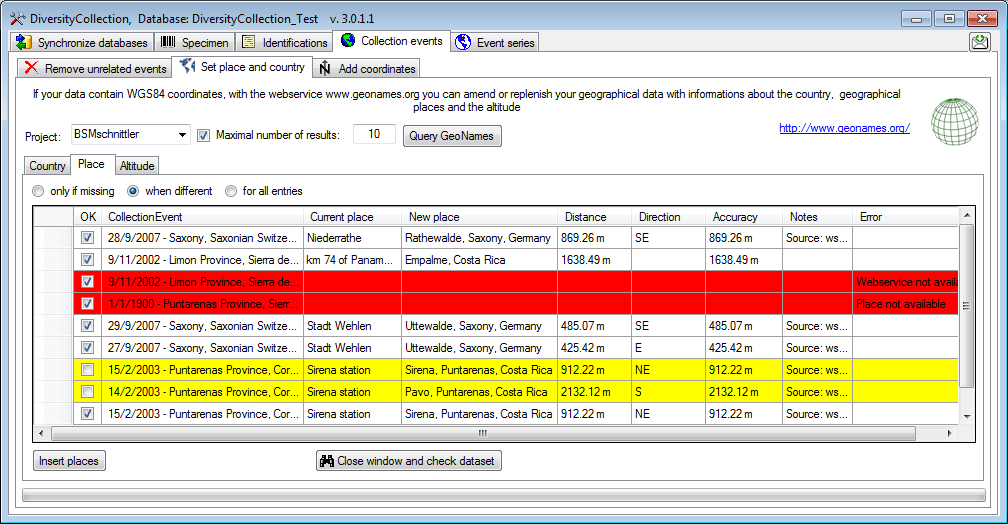
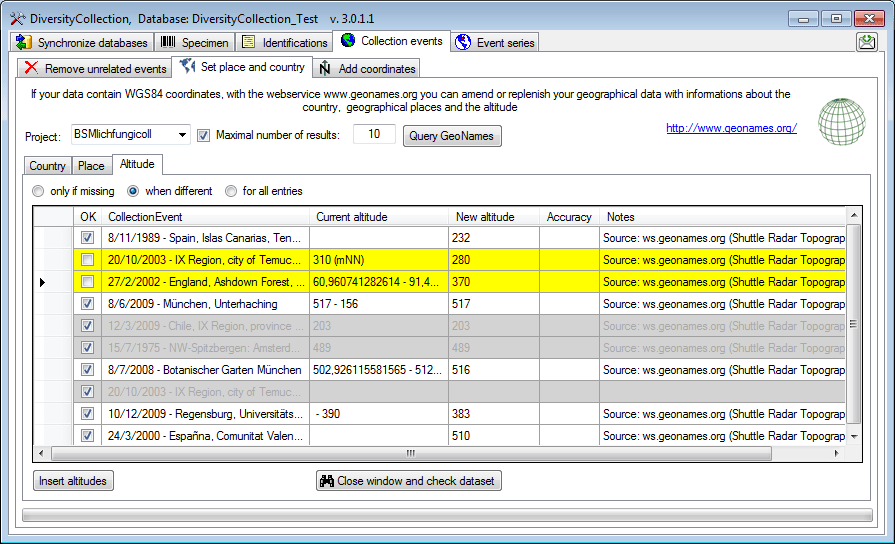
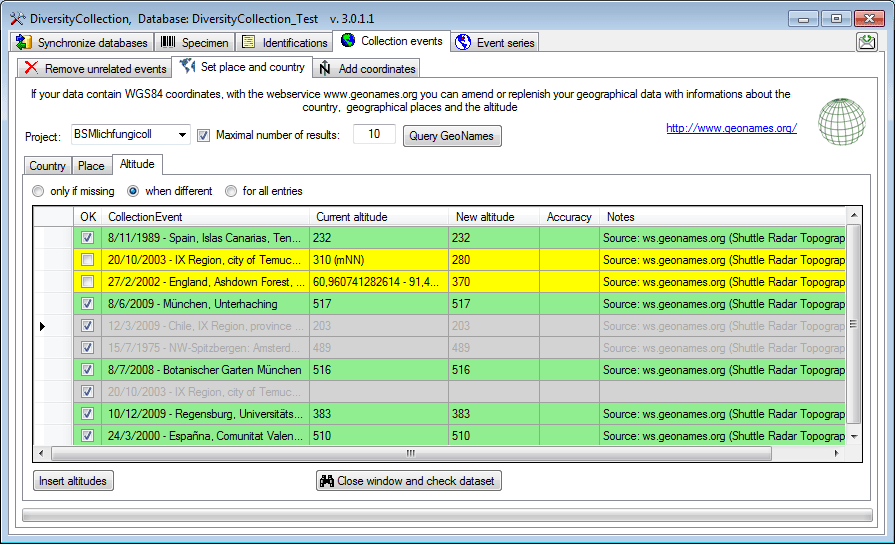
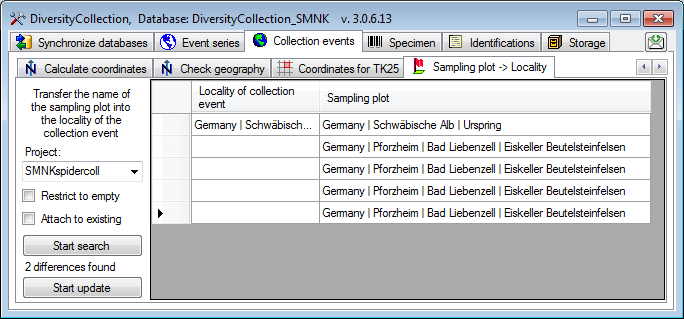
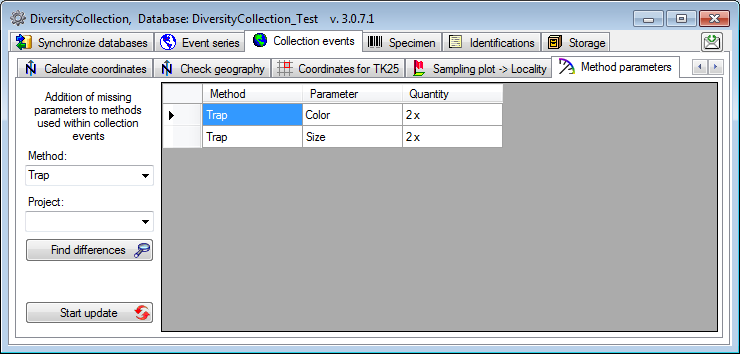
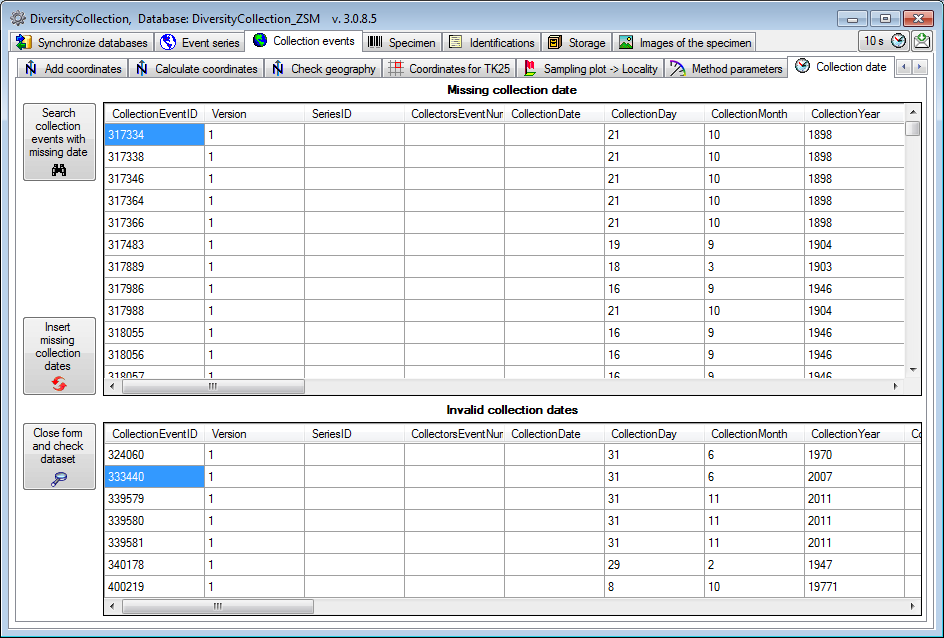
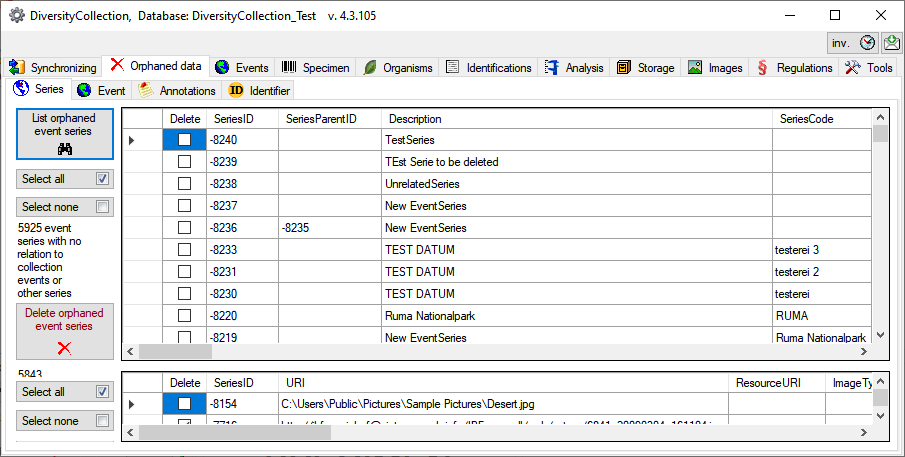
 button to start the
conversion. If a conversion is not possible, the fields for the target
system will be empty as shown below. The option
button to start the
conversion. If a conversion is not possible, the fields for the target
system will be empty as shown below. The option 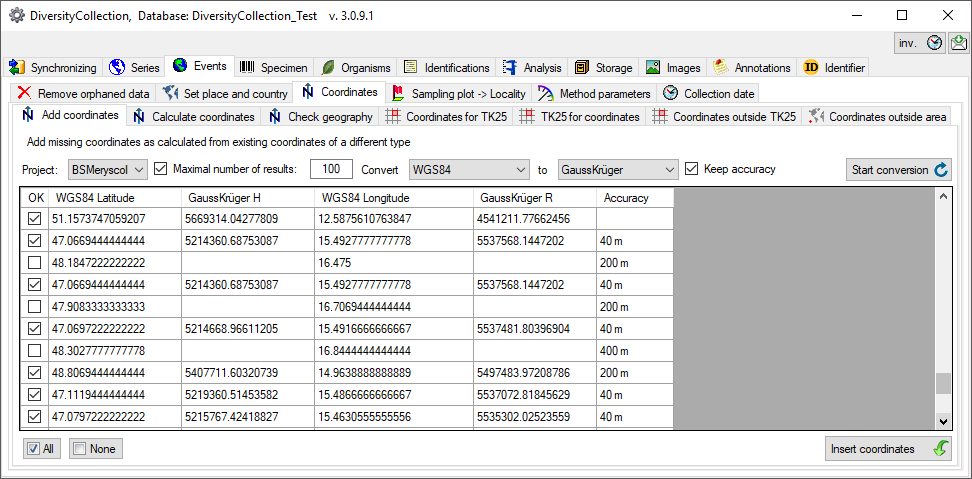
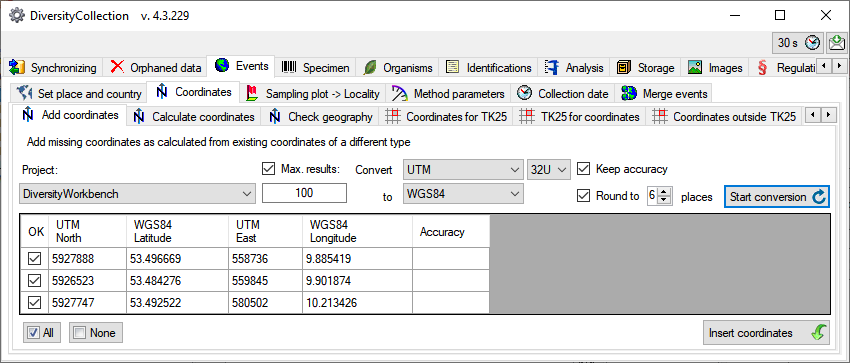
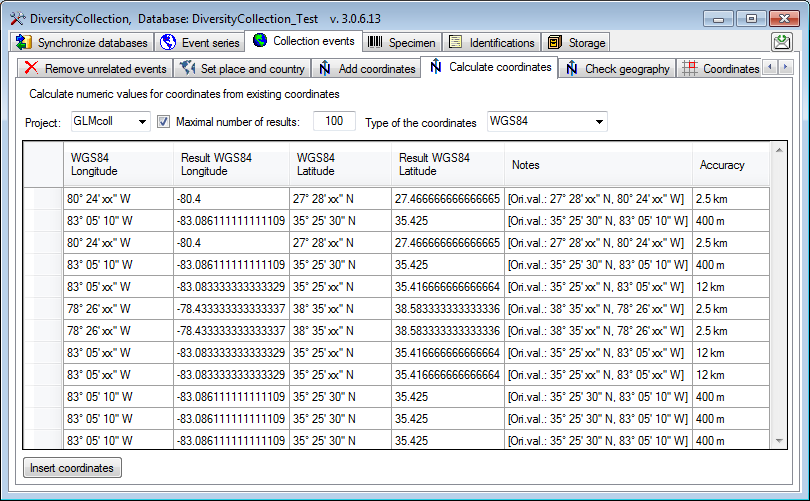
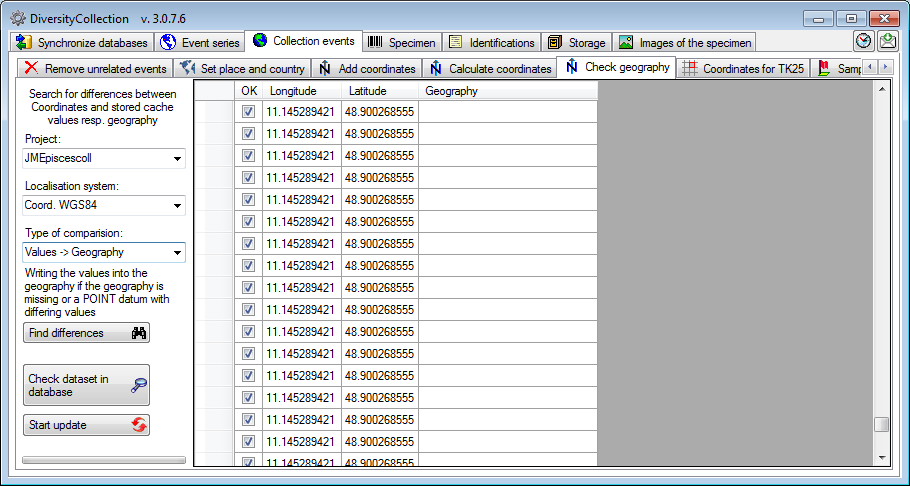
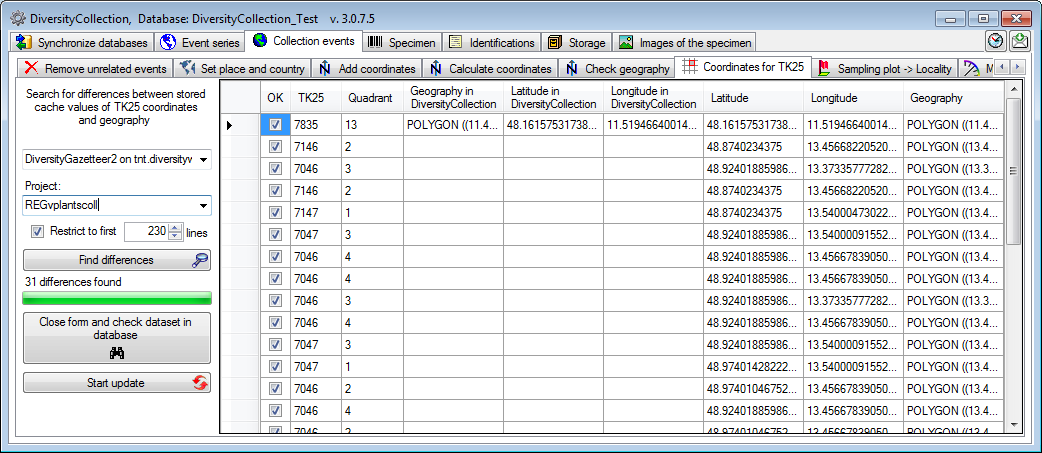
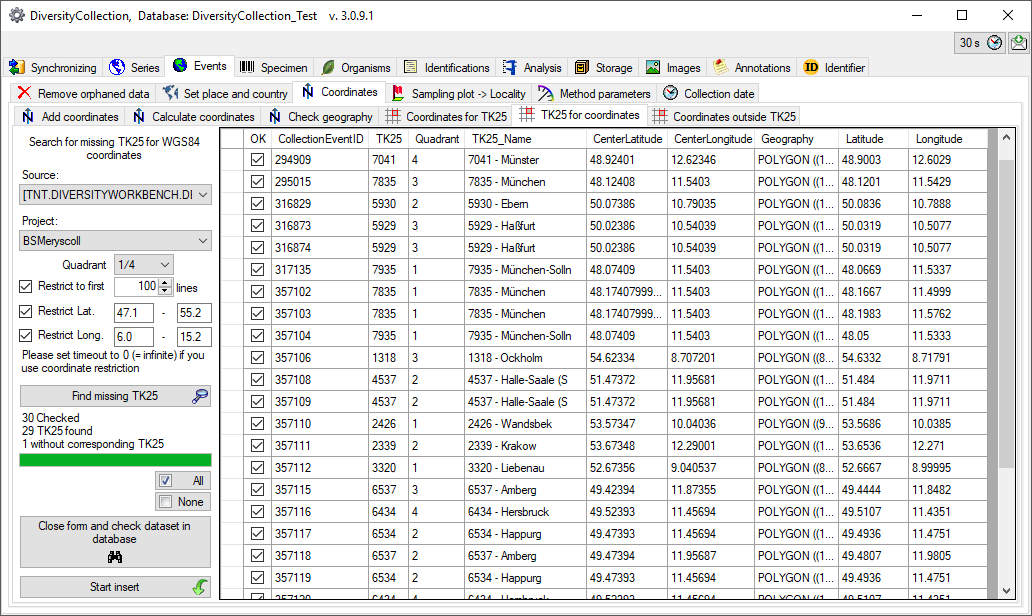
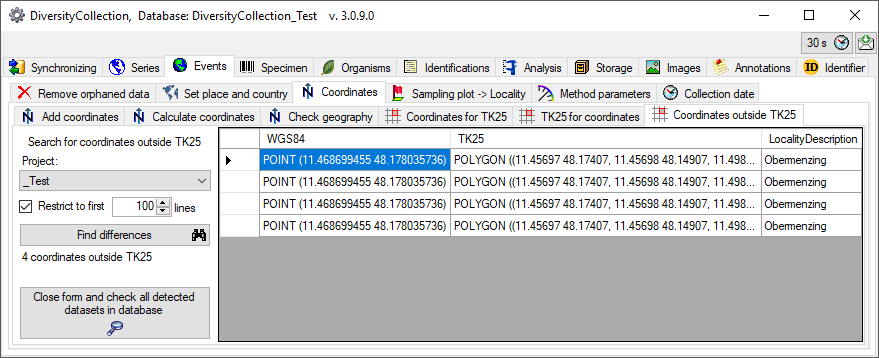
 outside an area
e.g. due to swapping latitude and longitude. You can detect these errors
by comparing all coordinates within a project with an area as provided
by DiversityGazetteer. In the maintenance form choose
outside an area
e.g. due to swapping latitude and longitude. You can detect these errors
by comparing all coordinates within a project with an area as provided
by DiversityGazetteer. In the maintenance form choose
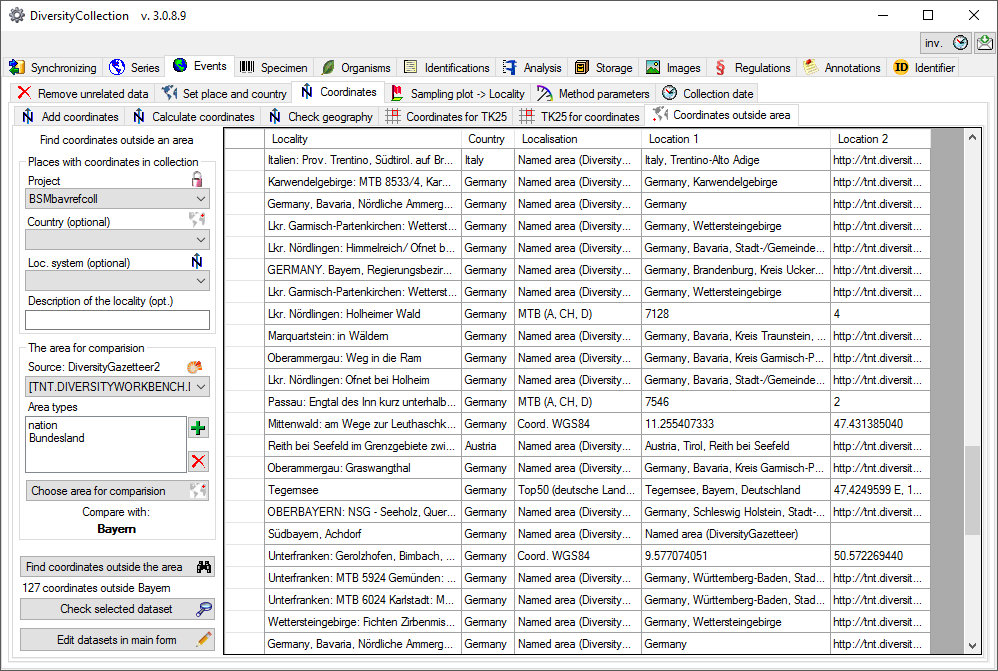
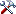 methods and images are included. To find
similar events with different entries in certain parts, you can deselect
those parts where there is no match in the data. To start the query
select a project and click on the
methods and images are included. To find
similar events with different entries in certain parts, you can deselect
those parts where there is no match in the data. To start the query
select a project and click on the 
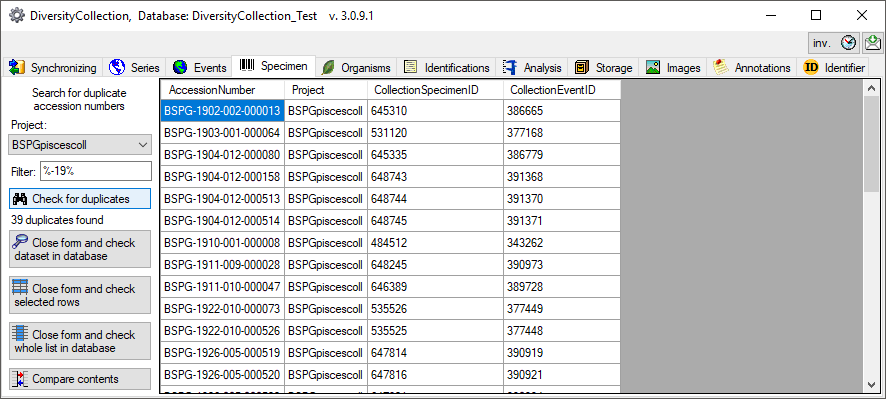
 . To list all duplicates in the main form
click on the Close form and check whole list in database
. To list all duplicates in the main form
click on the Close form and check whole list in database
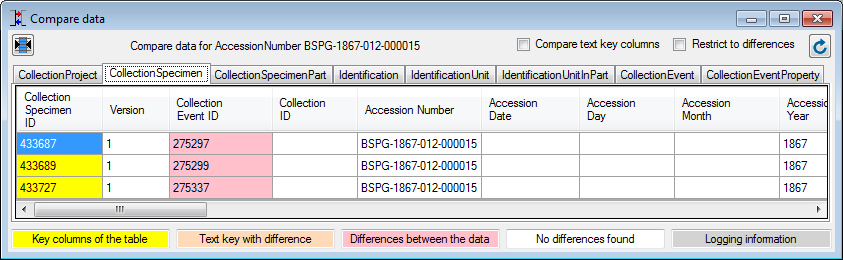
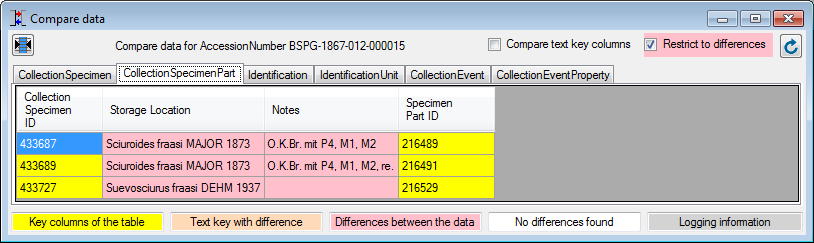
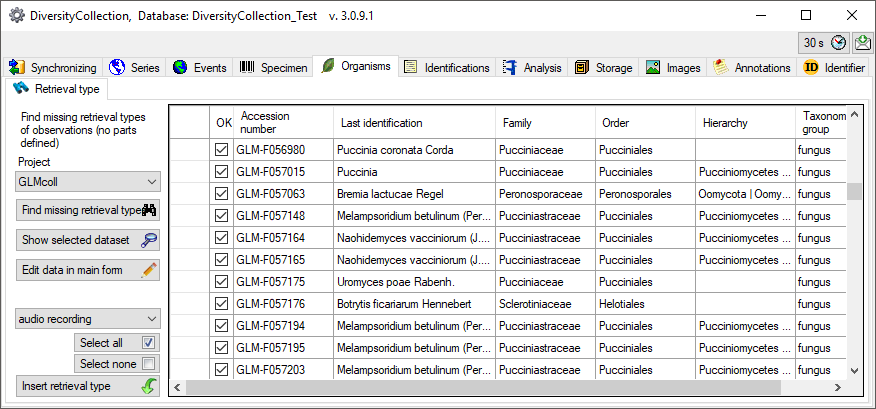
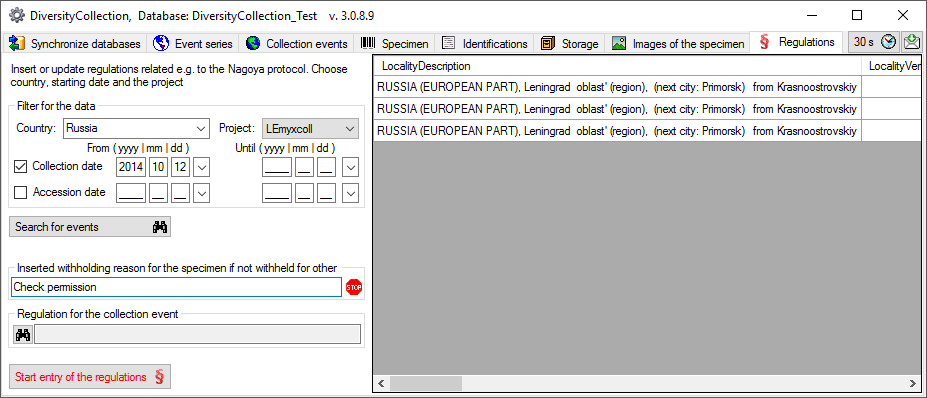

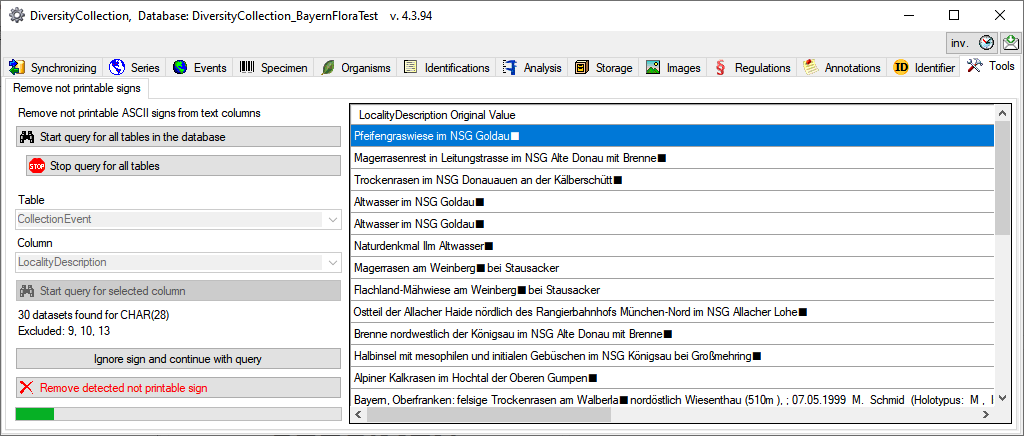
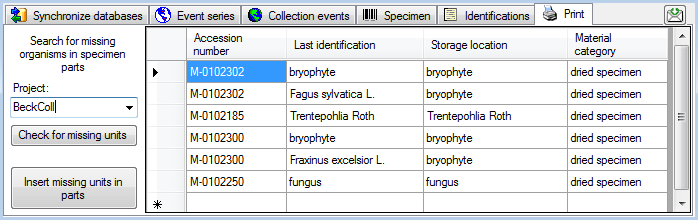
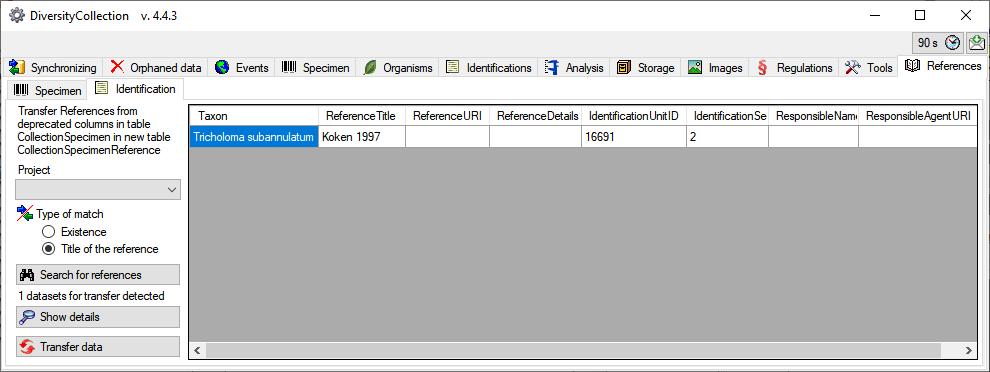





 ProjectAnalysis
ProjectAnalysis


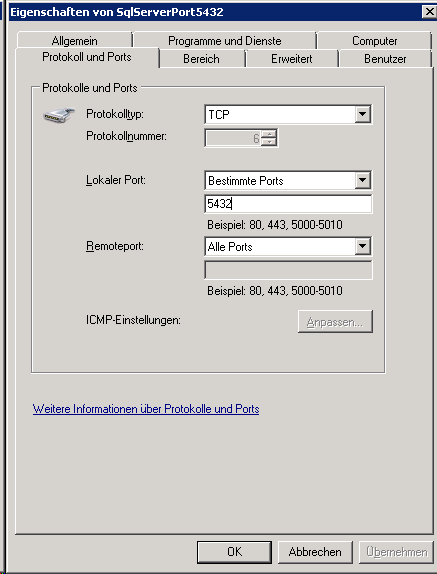
 windows login on the server and sysadmin permissions.
windows login on the server and sysadmin permissions.
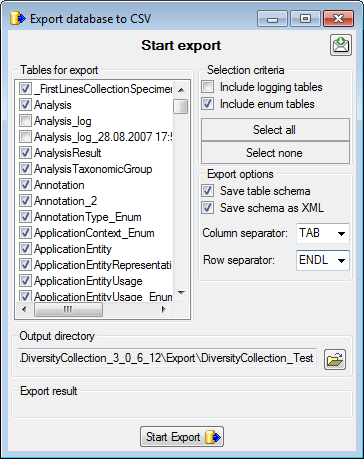











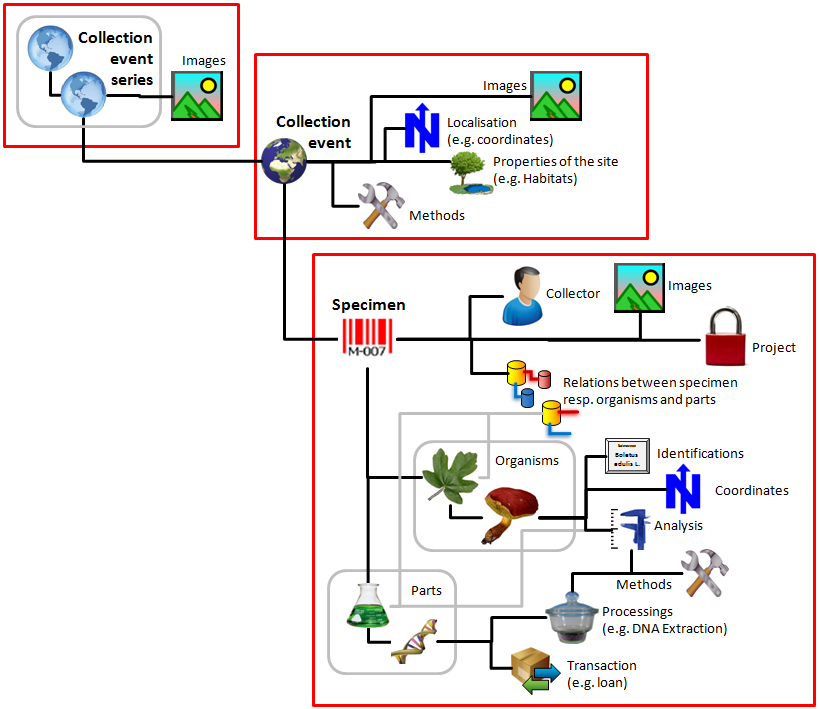
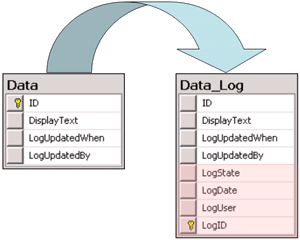





 button.
button. , uncheck
the enabled checkbox (see below).
, uncheck
the enabled checkbox (see below).





 button. A window a shown below will
open. It lists all
button. A window a shown below will
open. It lists all 
 button to copy the settings for all databases or the
button to copy the settings for all databases or the
 button. A window a shown below will open.
It lists all
button. A window a shown below will open.
It lists all 
 button may appear. This indicates, that
there are windows logins listed where the name of the login does not
match the logins of the server. This may happen if e.g. a database was
moved from one server to another. To correct this, click on the button.
A list of deviating logins will be shown, that can be corrected
automatically.
button may appear. This indicates, that
there are windows logins listed where the name of the login does not
match the logins of the server. This may happen if e.g. a database was
moved from one server to another. To correct this, click on the button.
A list of deviating logins will be shown, that can be corrected
automatically.









 General Data Protection Regulation of the European
Union several steps have to be performed in a database:
General Data Protection Regulation of the European
Union several steps have to be performed in a database:

 button (see below).
button (see below).




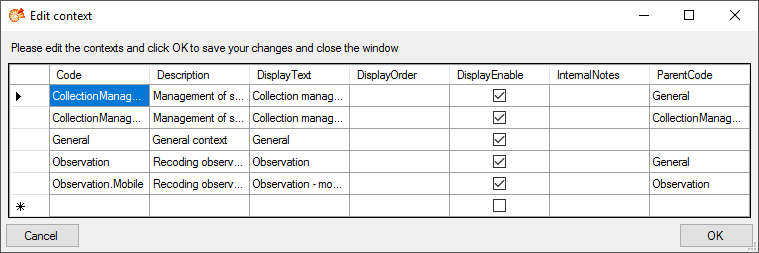



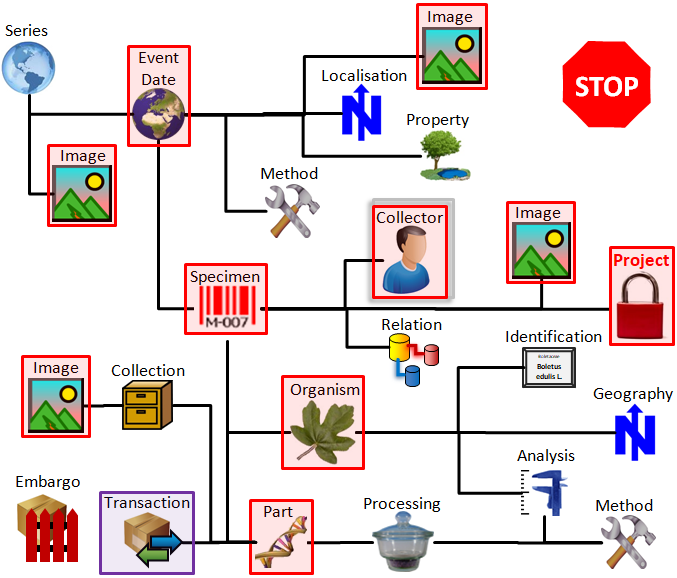
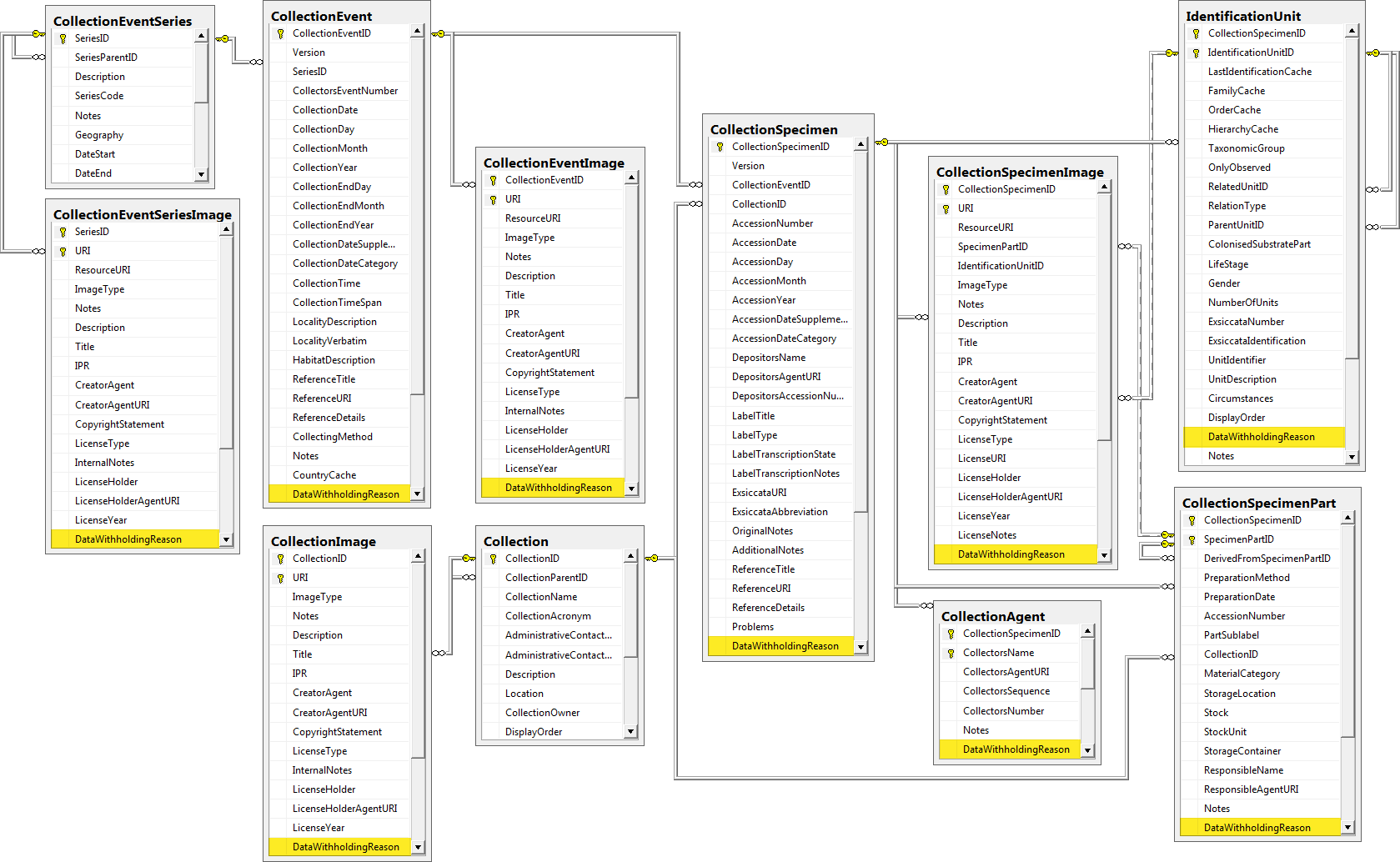
 Remove withholding reasons for all
... button or the
Remove withholding reasons for all
... button or the
 ... only from selected button, if only the
selected entry should be published.
... only from selected button, if only the
selected entry should be published.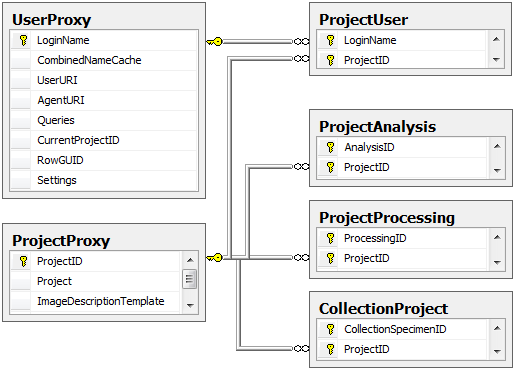

 Details of the projects within the DiversityWorkbench are stored in the
database DiversityProjects. To access further information on a project
click on the button. To edit details in projects you require the
application DiversityProjects.exe in your application directory and
access to the database DiversityProjects. To synchronize the projects
listed in DiversityProjects you may use the synchronize function in the
Details of the projects within the DiversityWorkbench are stored in the
database DiversityProjects. To access further information on a project
click on the button. To edit details in projects you require the
application DiversityProjects.exe in your application directory and
access to the database DiversityProjects. To synchronize the projects
listed in DiversityProjects you may use the synchronize function in the

 Clear ErrorLog.
Clear ErrorLog.
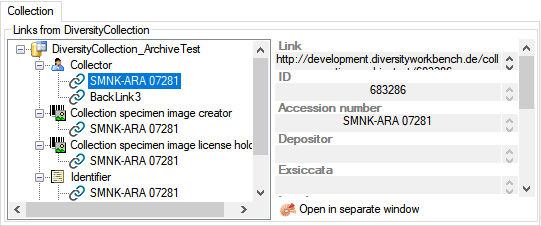
 Remote database
Remote database




 button to search the
webservice for matching entries. Choose among the provided results to
fill the textfield and the set the link to the webservice.
button to search the
webservice for matching entries. Choose among the provided results to
fill the textfield and the set the link to the webservice.