 DiversityCollection
DiversityCollection 
DiversityCollection (Version 3) is part of the database framework DiversityWorkbench. Within this framework the application DiversityCollection is confined to the management of specimens in scientific collections. In this context it is designed to document any action concerning the collection, storage, exchange and treatment of specimens in a collection and is also appropriate to store observation data. DiversityCollection is distinguished from other collection management systems by its focus on biological relations between organisms collected together as one or more specimens (e.g. host, parasite and hyperparasite, symbionts etc.). Any module within the Diversity Workbench is focused on a specific data domain. DiversityCollection keeps only data connected with the handling of collection specimens and observations. Data of other realms like e.g. taxonomy are handled in separate modules. For an overview of the available modules see DiversityWorkbench. DiversityCollection might also be used as a stand-alone application.
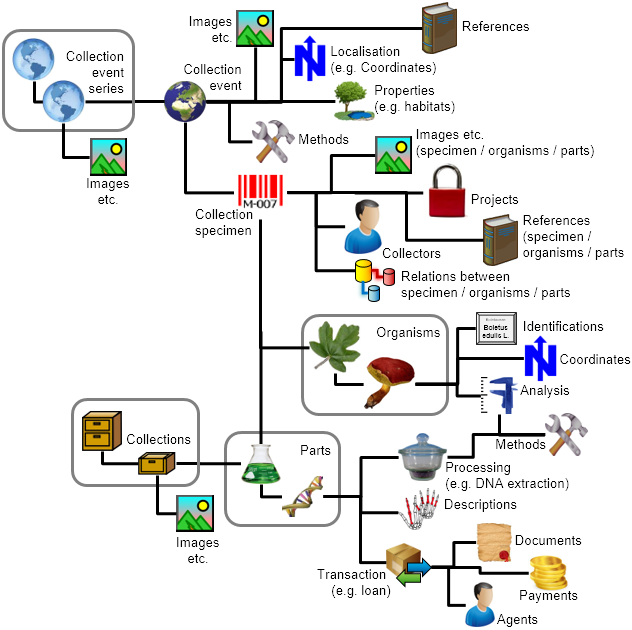
The image below gives you an overview of the main parts of DiversityCollection. The 4 grey squares in the image indicate the possibility of a hierarchy. This means that for example one organism can grow on another or a collection can be part of another collection.
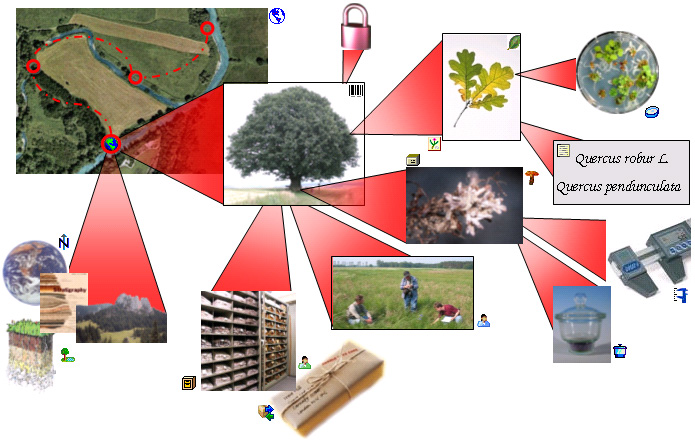
The image below gives an overview
of some typical data
depicted in DiversityCollection together with the symbols
used throughout the program.
A typical specimen
 may have been collected at a CollectionEvent
may have been collected at a CollectionEvent
 during an expedition
during an expedition
 . The site of the
CollectionEvent may
be localized
. The site of the
CollectionEvent may
be localized
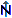 and characterized
and characterized
 and methods
and methods
 have been used.
The collectors
have been used.
The collectors
 collected twigs of the plant
collected twigs of the plant
 and fungi
and fungi
 growing on the plants. These organisms were identified
growing on the plants. These organisms were identified
 several times and analysed
several times and analysed
 where methods
where methods
 have been used. The collected samples are stored as herbarium sheets
have been used. The collected samples are stored as herbarium sheets
 and specimens
and specimens
 in a collection
in a collection
 . The manager
. The manager
 sent some of the samples
sent some of the samples
 for which he had a
request
for which he had a
request
 from a requester
from a requester
 to another collection. The samples were collected as part of
a project
to another collection. The samples were collected as part of
a project
 . Certain parts were
cultivated
. Certain parts were
cultivated
 and DNA
and DNA
 extracted which was processed
extracted which was processed
 where methods
where methods
 have been used. Images
have been used. Images
 were taken for the EventSeries, the CollectionEvent, the
CollectionSpecimen as
well as organisms and part of this specimen.
were taken for the EventSeries, the CollectionEvent, the
CollectionSpecimen as
well as organisms and part of this specimen.
DiversityCollection 4.x is based on Microsoft SQL-Server 2008 or later and the .Net Framework, Version 4.8.
For licence and copyright see the licence section.